Discovering New Restriction Enzymes
Script
So, the restriction enzymes are found in bacteria or archaea, but not higher organisms. So, you want to get some bacteria. You can go anywhere. So I mean, I've done them from my brother's backyard, one of my best enzymes. I was out visiting and I just took a water sample. I actually borrowed a little container from my sister in law, took a water sample with some mud in it, brought it back and tried to pick different looking colonies. You pick the small one, the big one, the white one, the yellow one, and I get maybe 20 different organisms that look different on a plate. And then you just get pure cultures.
So, you do basic microbiology. And then grill them up, break them open and say mix them with DNA. And do they have an enzyme that cuts DNA and gives you specific pieces? If they do, it's a type two restriction enzyme. So, then the fun begins, just do a little purification. And then it's kind of like doing a puzzle, which I love.
What we've done is we put a solution of our protein that we want to purify that still has some other things with it. So, we want to separate our protein of interest away from the other stuff. So in this case, we put five mLs of our sample of protein into this little tube. The pumps from the machine drive it through the tubing over onto the top of this column. It goes into the column and we keep running a buffer solution continually through it. And the proteins actually migrate their way down through the column. So, the solution is going to come down and then come over, and we're actually collecting fractions this machine, has a bunch of individual tubes for fractions and it's going to collect a certain volume and then move to the next tube and then collect the same volume and keep going.
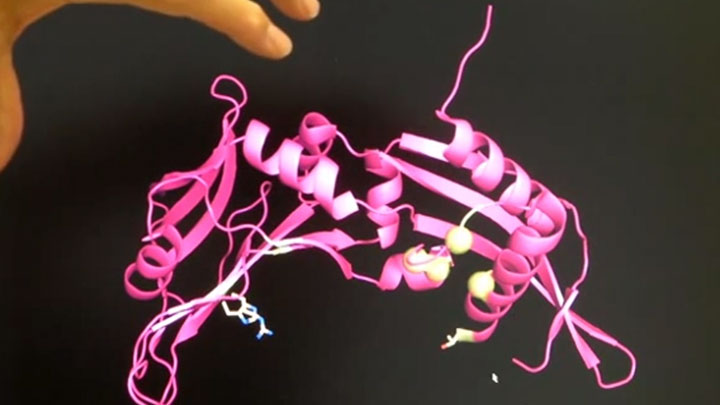
Here we can see a trace of what's been happening. So the blue line is the UV absorbance. So, it's going to... when the proteins come off, you'll get an increase in absorbance as they go past the detector. So there's basically nothing happening. And this is the protein we're after, gave us a nice peak and you don't see a whole lot else because this is already fairly pure. So, this is actually looking really good. I'm happy with this. So, I'll send this down to New York. They'll mix it with DNA, hopefully get it to form crystals, and then they can trace how the protein is actually structured.
When you cut DNA, so you cut a few different DNAs or just one, they'll give you different size pieces. You can basically take some DNA, cut it, run a gel to separate things by size. And then look at it and say, "Okay, I have a different pattern of sizes. They're different enzymes." What we would like is to find new ones. So, things that cut at a new site, or cut even the same sequence a different way. It just, then you have more tools to use.
This is a picture of an agarose gel. So, you can see up here they're the wells with the holes in the gel. And I've put the DNA solution into these, which sit down into the gel and then run a current from the top to the bottom. And the DNA migrates through the agarose, and it's kind of like people. The small pieces of DNA can work their way through the gel much faster than the big pieces of DNA. So, you separate things based on size. So, these are the largest pieces. They moved the shortest distance from here and these are the smallest pieces. They've moved along... significantly a further distance.
So in this case, this particular one here is a DNA size standard. So these are known pieces of DNA that I know the sizes of. So, that tells me where I am in the gel on this particular run. So, it's a control and it tells you... I can compare this to my known piece in this known piece is 1,274 base pairs. So, that's a little bit bigger. That's maybe 1280, 1290, 1300, something like that.
The question I'm answering here is how much enzyme do I have? This is lambda DNA, which is a one big intact piece. And it's cut with different amounts of enzyme. And we're looking at how much enzyme do I need to cut this into the... at every place where this enzyme recognizes the DNA. So, it would be a complete digest. So, this would be... These are complete digest. These are clearly incomplete. This is the highest concentration to lowest.
Related Videos
-

What are Enzymes for Innovation? -
How do restriction enzymes interact with DNA? -

What are Restriction Enzymes?

