Choosing Input Amounts for the Monarch HMW DNA Extraction Kits
Cells
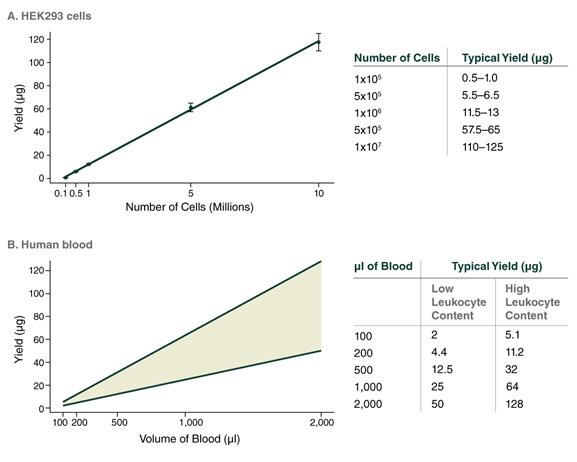
The sample input range is 1 x 105–1 x 107 cells, but an input amount of 1 x 106 cells is recommended. The upper limit for cell input amounts is dictated by the viscosity of the lysed sample, which poses a challenge to the efficiency of the enzymes, precipitation onto the beads, and the dissolving of the purified DNA. If using more than 2 x 106 cells, purified DNA samples will be viscous and may be more difficult to dissolve and handle. If using an input below 1 x 105 cells, DNA recovery will be significantly reduced. It is important to note that if employing a low agitation speed during lysis, inputs should not exceed 5 x 106 cells. When working with samples less than 5 x 105, follow protocol guidance for Low Input to ensure the buffer volumes used reflect the lower cell count.
Blood
Mammalian Blood
The sample input range for mammalian blood samples is 100 µl up to 2 ml. Working with a starting sample of 500 µl is recommended for most sample types; 200 µl is recommended for rabbit samples, which have a high cell content. If working with starting volumes > 500 µl, samples will need to be initially processed in larger tubes or split into 500 µl aliquots. The container needs to accommodate the addition of 3 volumes of RBC Lysis Buffer; working with 500 µl enables processing in a 2 ml tube. If splitting the sample into multiple aliquots, the pellets can be combined into a single 2 ml tube after RBC lysis to facilitate further processing. If working with sample volumes below the recommended input amount (500 µl for most samples), follow the protocol guidance for Low Input to ensure the buffer volumes reflect the lower cell count.
Nucleated Blood
The maximum input for successful DNA extraction from nucleated blood is 20 µl; exceeding this will overload the system. The recommended input amount is 5 µl.
Table 1: Guidance on sample input amounts and expected results: cells & blood (NEB #T3050)
Table 1 provides data on minimum, maximum, and recommended input amounts for various cell lines and blood samples using the Monarch HMW DNA Extraction Kit for Cells & Blood. Data on yield, purity, and RNA content is also provided. Samples that were successfully tested in standard ligation-based Oxford Nanopore Technologies sequencing runs are indicated. RNA content was determined by HPLC analysis of nucleoside content after digestion of 1 µg of eluted nucleic acid with the Nucleoside Digestion Mix (NEB #M0649). Yields from blood samples vary by donor due to different leukocyte content; yield can vary up to 3-fold by donor. Similar yield and purity results were obtained with different anticoagulants (e.g., EDTA, citrate, heparin and PAXgene Blood DNA tubes were tested).
Using input amounts below the recommended minimum will reduce yields drastically. Exceeding maximum input amounts will result in DNA eluates that are highly viscous and difficult to dissolve and will reduce purity of the isolated DNA. Results are shown for samples that were lysed with agitation at 2,000 rpm.
|
MINIMUM INPUT (CELLS) |
MAXIMUM INPUT (CELLS)* |
RECOMMENDED INPUT AMOUNT (CELLS) |
YIELD (μg) FROM 1 x 106 CELLS |
PURITY RATIOS |
RNA CONTENT |
VALIDATED FOR ONT SEQUENCING? |
|||
|---|---|---|---|---|---|---|---|---|---|
|
A260/280 |
A260/230 |
||||||||
|
HEK293 |
1 x 105 |
1 x 107 |
1 x 106 |
11.5–13 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
|
HeLa |
1 x 105 |
1 x 107 |
1 x 106 |
12.9 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
|
NIH3T3 |
1 x 105 |
1 x 107 |
1 x 106 |
9.4 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
|
Jurkat |
1 x 105 |
1 x 107 |
1 x 106 |
13.7 |
1.86 |
2.5 |
≤ 1% |
Yes |
|
|
K562 (suspension cells) |
1 x 105 |
1 x 107 |
1 x 106 |
13.7 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
|
HCT116 |
1 x 105 |
1 x 107 |
1 x 106 |
16.9 |
1.86 |
2.5 |
≤ 1% |
Yes |
|
|
A549 |
1 x 105 |
1 x 107 |
1 x 106 |
12.7 |
1.86 |
2.3 |
≤ 1% |
Yes |
|
|
U5Os |
1 x 105 |
1 x 107 |
1 x 106 |
10.6 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
|
HepG2 |
1 x 105 |
1 x 107 |
1 x 106 |
13.4 |
1.81 |
2.2 |
≤ 1% |
Yes |
|
|
NCI-460 |
1 x 105 |
1 x 107 |
1 x 106 |
9.5 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
|
SK-N-SH |
1 x 105 |
1 x 107 |
1 x 106 |
9.5 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
|
Aa23 |
1 x 105 |
1 x 107 |
1 x 106 |
8.7 |
1.81 |
2.3 |
≤ 1% |
Yes |
|
|
Mammalian Blood |
|||||||||
|
MINIMUM INPUT (μl) |
MAXIMUM INPUT (μl)* |
RECOMMENDED INPUT AMOUNT (μl) |
YIELD (μg) for 500 µl** |
PURITY RATIOS |
RNA CONTENT |
VALIDATED FOR ONT SEQUENCING? |
|||
|
A260/280 |
A260/230 |
||||||||
|
Human*** |
Fresh |
100 |
2,000 |
500 |
12–32 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
Frozen |
100 |
2,000 |
500 |
9–30 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
|
Mouse |
Fresh |
100 |
2,000 |
500 |
7–11 |
1.88 |
2.4 |
≤ 1% |
Yes |
|
Frozen |
100 |
2,000 |
500 |
16–17 |
1.88 |
2.4 |
≤ 1% |
ND |
|
|
Rat (fresh only) |
Fresh |
100 |
2,000 |
500 |
29–38 |
1.87 |
2.4 |
≤ 1% |
Yes |
|
Rabbit |
Fresh |
100 |
500 |
200 |
12–15 |
1.72 |
1.9 |
≤ 1% |
Yes |
|
Fresh |
100 |
500 |
200 |
200 µl: 4–5 |
1.89 |
2.4 |
≤ 1% |
Yes |
|
|
Frozen |
100 |
500 |
200 |
200 µl: 4–5 |
1.89 |
2.4 |
≤ 1% |
Yes |
|
|
Pig |
Fresh |
100 |
2,000 |
500 |
up to 42 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
Frozen |
100 |
2,000 |
500 |
up to 40 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
|
Horse |
Fresh |
100 |
2,000 |
500 |
16 |
1.86 |
2.3 |
≤ 1% |
Yes |
|
Frozen |
100 |
2,000 |
500 |
22.3 |
1.86 |
2.4 |
ND |
ND |
|
|
Cow |
Fresh |
200 |
2,000 |
500 |
7 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
Frozen |
200 |
2,000 |
500 |
9.1 |
1.86 |
2.4 |
ND |
ND |
|
|
Rhesus monkey |
Fresh |
100 |
2,000 |
500 |
52 |
1.86 |
2.4 |
≤ 1% |
Yes |
|
Frozen |
100 |
2,000 |
500 |
52.6 |
1.86 |
2.5 |
ND |
ND |
|
|
Goat (fresh only) |
Fresh |
100 |
2,000 |
500 |
24 |
1.87 |
2.4 |
≤ 1% |
Yes |
| Sheep (fresh only) |
Fresh | 100 | 2,000 | 500 | 15.3 | 1.87 | 2.4 | ND | ND |
|
Nucleated Blood |
|||||||||
|
MINIMUM INPUT (μl) |
MAXIMUM INPUT (μl)* |
RECOMMENDED INPUT AMOUNT (μl) |
YIELD (μg) per 5µl |
PURITY RATIOS |
RNA CONTENT |
VALIDATED FOR ONT SEQUENCING? |
|||
|
A260/280 |
A260/230 |
||||||||
|
Chicken |
Fresh |
2 |
20 |
5 |
33 |
1.86 |
2.5 |
ND |
Yes |
|
Frozen |
2 |
20 |
5 |
30 |
1.86 |
2.5 |
ND |
ND |
|
|
Turkey |
Fresh |
2 |
20 |
5 |
37 |
1.87 |
2.4 |
ND |
Yes |
|
Frozen |
2 |
20 |
5 |
28 |
1.87 |
2.5 |
ND |
ND |
|
ND = Not determined
* For low agitation speeds, do not exceed 5 x 106 cells
** Unless otherwise stated
*** Compatible with K2-EDTA, Na-citrate, Na-heparin, PAXgene® Blood DNA
Tissue
The sample input range is 2–25 mg for most tissues (2–15 mg of DNA-rich / soft organ tissues (e.g., kidney, liver), 2–25 mg for brain). The upper limit for tissue input amounts is often limited by the viscosity of the lysed sample, which negatively impacts enzyme access, protein removal, precipitation onto the beads, and dissolving/resuspension of the purified DNA. In some samples, the high amounts of fibers or fatty acids can be factors that limit the input amounts. If a lower-than-recommended input amount is used, DNA recovery will be significantly reduced. Standard and low input protocols are provided to ensure the buffer volumes are appropriate and that precipitation onto the beads is efficient. If working with fatty or fibrous tissues (e.g., brain and muscle), and only very small amounts of sample are available (< 5 mg), see guidance below.
Using Very Low Input Amounts
In some cases, inputs below the range provided can be successfully processed using this kit if the volume of lysate, reagents and buffers are reduced, as described below. For brain and muscle samples, input amounts of 2–5 mg have been successfully processed. When working with samples in this input range, stop agitation after 15 minutes during the 45-minute lysis for maximum yield.
- Part 1, Step 1: use 100 µl Tissue Lysis Buffer and 10 µl Proteinase K
- Part 1, Step 6: use 5 µl RNase A
- Part 1, Step 8: use 55 µl Protein Separation Solution
- Part 2, Step 2: use 90 µl isopropanol
Bacteria
The sample input range for E. coli is 5 x 108 – 5 x 109 cells. As described for tissue samples, the upper limit for bacteria input amounts is limited by the viscosity of the lysed sample, which negatively impacts enzyme access, protein removal, precipitation onto the beads, and dissolving/resuspension of the purified DNA. If an input amount below the recommended amount is used, DNA recovery will be significantly reduced. Standard and low input protocols are provided to ensure the buffer volumes are appropriate for the sample input amount used. The sample input range for B. cereus using the low input protocol is 2 x 108 – 4 x 108 cells.
Table 2: Guidance on sample input amounts and expected results: tissue, bacteria and other samples (NEB #T3060)
The table below provides guidance on the minimum, maximum, and recommended input amounts for various sample types when using the Monarch HMW DNA Extraction Kit for Tissue. Data on yield, purity, and RNA content is also provided. Samples that were successfully tested in standard ligation-based Oxford Nanopore Technologies sequencing runs are indicated. Using input amounts that exceed the maximum will lead to challenges in solubility and viscosity, and purity may be affected. If more starting material is required, splitting the sample and performing multiple preps is recommended. RNA content was determined by HPLC analysis of nucleoside content after digestion of 1 µg of eluted nucleic acid with the Nucleoside Digestion Mix (NEB #M0649). Using input amounts below the recommended minimums will reduce yields drastically.|
MINIMUM INPUT (mg) |
MAXIMUM INPUT (mg)* |
RECOMMENDED INPUT AMOUNT (mg) |
YIELD (µg) FOR RECOMMENDED INPUT (YIELD PER mg) |
PURITY RATIOS |
RNA CONTENT |
VALIDATED FOR ONT SEQUENCING? |
|||
|---|---|---|---|---|---|---|---|---|---|
|
A260/280 |
A260/230 |
||||||||
|
Mammalian Tissue |
|||||||||
|
Mouse brain |
Fresh |
2** |
20 |
15 |
12–21 |
1.87 |
2.39 |
ND |
YES |
|
Frozen |
2** |
20 |
15 |
15–21 (1–1.5) |
1.86 |
2.48 |
ND |
YES |
|
|
Mouse liver |
Fresh (w/NaCl) |
2 |
15 |
10 |
7 |
1.84 |
2.10 |
1.2% |
YES |
|
Frozen (w/NaCl) |
2 |
15 |
10 |
17–19 (1.7–1.9) |
1.89 |
2.50 |
ND |
YES |
|
|
Fresh* |
2 |
15 |
10 |
20 |
1.84 |
1.52+ |
8.7% |
YES |
|
|
Frozen* |
2 |
15 |
10 |
27–31 (2.7–3.1) |
1.89 |
1.93++ |
ND |
YES |
|
|
Mouse muscle |
Fresh |
2** |
25 |
20 |
8–9 |
1.87 |
2.25 |
2.1% |
YES |
|
Frozen |
2** |
25 |
20 |
12–16 (0.6–0.8) |
1.87 |
2.30 |
ND |
YES |
|
|
Mouse kidney |
Fresh |
2 |
15 |
10 |
23–34 |
1.86 |
2.44 |
ND |
YES |
|
Frozen |
2 |
15 |
10 |
32–41 (3.2–4.1) |
1.86 |
2.53 |
0.8% |
YES |
|
|
Mouse tail |
Frozen |
2** |
25 |
20 |
20 (1.8–2.1) |
1.86 |
2.43 |
ND |
YES+++ |
|
Mouse ear punch |
Fresh |
2** |
15 |
10 |
15–16 (1.5–1.6) |
1.86 |
2.29 |
ND |
YES |
|
Rat kidney |
Frozen |
2 |
15 |
10 |
20–25 |
1.87 |
2.40 |
ND |
YES |
|
Bacteria |
|||||||||
|
E. coli |
Frozen |
5 x 108 cells |
5 x 109 cells |
1 x 109 cells |
8–9 |
1.89 |
2.31 |
1.7% |
YES |
|
B. cereus |
Frozen |
2 x 108 cells |
4 x 108 cells |
2 x 108 cells |
4–5 |
1.86 |
2.20 |
3.9% |
YES |
|
M. luteus |
Frozen |
ND |
ND |
1 x 108 cells |
2.0 |
1.89 |
2.09 |
ND |
ND |
|
Amphibian |
|||||||||
|
X. laevis |
Fresh |
ND |
ND |
3–4 |
5 |
1.86 |
2.51 |
2.3% |
ND |
|
Yeast |
|||||||||
|
S. cerevisiae |
Fresh |
ND |
ND |
20 x 107 cells |
3–6 *** |
1.90 |
2.01 |
ND |
ND |
|
Insect |
|||||||||
|
A. aegypti |
Frozen |
ND |
ND |
15 |
6 |
1.84 |
2.53++ |
2.7% |
ND |
|
Nematodes |
|||||||||
| C. elegans **** |
Frozen | ND | ND | 2 plates | 8.2 | 1.91 | 2.5 | ND | ND |
ND = Not determined
* Standard protocol without recommended NaCl treatment
** If working with input amounts <5 mg, refer to the product manual for guidance on reducing buffer volumes
*** Total nucleic acid yields are 4-10 µg and 6-12 µg for haploid and diploid strains, respectively. Though an RNase A step is included, RNA is co-purified. Yields may vary depending on the strain.
**** Rotor-stator homogenization is recommended
+ Measured with Nanodrop One; systems that differentiate turbidity in the content profiling will give higher values
++ Measured with Unchained Labs Lunatic (formerly Trinean DropSense16); devices without content profiling that differentiates turbidity may give lower values
+++ Size selection is recommended.
Related Products:
Related Resources:
- Monarch HMW DNA Extraction Kit for Cells & Blood Product Manual
- Monarch HMW DNA Extraction Kit for Tissue Product Manual
- Choosing an Agitation Speed During Lysis with the Monarch HMW DNA Extraction Kits
- Homogenization of High Molecular Weight DNA (HMW DNA) Samples
- Measuring, Analyzing & Storing High Molecular Weight DNA (HMW DNA) Samples
- Troubleshooting Guide for High Molecular Weight DNA Extraction Using the Monarch HMW DNA Extraction Kits
- A faster workflow for the assessment of genomic loci in mice using a novel HMW DNA extraction technology upstream of Cas9 targeted sequencing
- ’ Ultra-Long DNA Sequencing Workflow®Guidance for HMW DNA Extraction Upstream of Oxford Nanopore Technologies

