Tips for successful RNA Cleanup using the Monarch® RNA Cleanup Kits
Script
Successful RNA purification using the Monarch Total RNA Mini Prep Kit can easily be accomplished by following these helpful tips.
Part 1 Before Your Prep
1.1 Eliminate RNases From the Work Environment
Before your prep. It is important to work in an environment free from RNases. Always wear gloves, use RNase-free glass and plastic wear and clean your work areas. We recommend wiping your bench top with a cleaning agent such as RNaseZAP.
1.2 Store RNA Samples Properly
To maintain RNA integrity of your sample, prior to the prep, samples should be frozen at the time of harvest, or protected by a dedicated reagent such as the Monarch DNA and RNA Protection Reagent, which is included in our kit. Cultured cell pellets are aliquots of blood preserved with this protection reagent, should be mixed thoroughly by vortexing or pipetting prior to storage.
Small pieces of tissue less than 20mg may be stored directly in the protection reagent. However, larger tissue samples should be homogenized in protection reagent prior to storage. If samples have been preserved in the protection reagent, do not remove the liquid prior to processing, as the protection reagent also begins the lysis of the sample.
1.3 Avoid Thawing Samples
It's important to avoid thawing your sample in order to preserve it's integrity before extraction of the RNA. Frozen cultured cells can be thawed briefly before the addition of the RNA lysis buffer, but most other sample types should not be thawed in the absence of the protection reagent or the lysis buffer. These samples include tissues, blood, bacteria, yeast and plants.
1.4 Maximize Your Yield by Ensuring Complete Homogenization
Ensure that your samples are completely disrupted and homogenized in order to release all of the RNA and maximize your yield. Mechanical homogenization for example, with a bead homogenizer, can boost your yields beyond those produced by proteinase K digestion alone when you're working with tissue. Mechanical homogenization is also recommended for tough to lyse samples such as plant, bacteria, and yeast.
1.5 Multiple Rounds of Homogenization May Be Recommended
When using mechanical lysis and homogenization, multiple rounds of homogenization may be recommended. If you do use multiple rounds, place your sample on ice for approximately one minute in between rounds to prevent your sample from over heating.
1.6 For Tissues or Leukocytes Preheat the heating block
If you are working with tissues or leukocytes it can help to preheat your heating block to 55 degrees Celsius before starting your prep, so that the heat block will be at the correct temperature when you get to the proteinase K incubation. Please be aware that when working with mammalian whole blood, the proteinase K incubation is carried out at room temperature.
1.7 Prepare Master Mix
We also recommend preparing a master mix of DNase I and DNase I reaction buffer prior to beginning if you are performing multiple preps at once.
Part 2 During Your Prep
2.1 Follow NEB Guidance
Be sure to follow our guidance on the recommended sample input amounts in order to ensure that the buffer volumes are appropriate and that the columns are not overloaded. This is a common mistake that quickly can reduce yield, purity, and integrity of your RNA.
2.2 After Lysis, Perform Procedure at Room Temp
After sample lysis, perform all steps at room temperature. This will prevent detergent from precipitating in the buffers. Do not place your samples on ice after lysis.
2.3 Label Collection Tubes
Before applying your sample to the genomic DNA removal column, be sure that you label the collection tube, as you'll be discarding the column after you centrifuge. After putting your sample through the genomic DNA removal column, make sure that you've saved the flow through. This flow through contains your RNA.
2.4 Add One Volume of Ethanol to Your Flow-Through
If you want to capture total RNA including small RNAs, add one volume of ethanol to your flow through from the gDNA removal column, in preparation for binding it to the RNA purification column. If you want to exclude RNAs smaller than two hundred nucleotides, only add half the volume of ethanol to your flow through from the gDNA removal column.
2.5 Perform All Wash Steps and Spin for Two Minutes after Your Last Wash
It's very important to perform all wash steps in the protocol in order to produce highly pure RNA. Be sure to spin your column for two minutes after the last wash.
2.6 For Low Yield Samples, Elute in 50ul Nuclease Free Water
For low yield samples, including muscle or brain, or for low starting inputs, consider eluting the RNA with 50 microliters of nuclease free water in order to get concentrated RNA. It's important to have a concentration above 20 nanograms per microliter. If a microvolume spectrophotometer, such as a NanoDrop, will be used to measure concentration and purity.
Part 3 After Your Prep
3.1 Spin to Remove Silica
In some cases, silica particles from the column matrix can be found in your eluate. To ensure that this doesn't affect your OD260-230 ratio, you can centrifuge the eluate for one to two minutes at 1600 x G and pipette the aliquot from the time of the liquid for measurement on a spectrophotometer.
3.2 Add EDTA to Protect RNA and Store in Aliquots
While nucleus free water is provided in this kit for the elution of RNA, adding EDTA to a final concentration of .1 to 1 millimolar can protect RNA samples that will be stored for an extended period of time. Additionally, it's good practice to store the eluted RNA in aliquots to avoid excessive freeze-thaw cycles.
Contact our Technical Support
We hope that these tips have been helpful. If you have any further questions, our tech support staff would be happy to help. Contact us at info@neb.com or online at neb.com/tech support.
Related Videos
-

Tips for Successful RNA Purification using Monarch® Total RNA Miniprep Kit -
Monarch® RNA Cleanup Protocol -
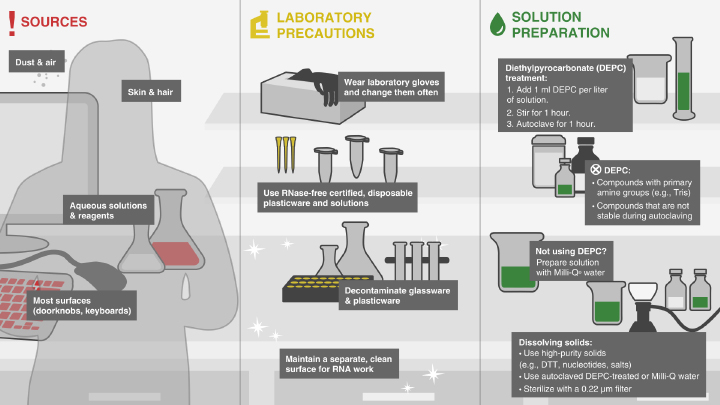
Avoiding RNase Contamination

