Automating and miniaturizing NEB workflows for high-throughput DNA assembly and protein expression

Script
Jackson Buss:
Hello and welcome. My name is Jackson Buss, and I'm a Research Scientist at New England Biolabs. My main focus here is to develop and employ high-throughput screening tools to identify new and better enzymes. Since I exclusively use NEB products in my workflows, I've learned to use them effectively and efficiently. Here I hope to convey this knowledge to you as I described the automation and miniaturization of NEB workflows for high-throughput DNA assembly and protein expression.
In this talk, we will cover how to perform high-throughput plasmid construction using NEBridge Golden Gate Assembly, and I will also cover how to perform multi-site directed mutagenesis using NEBuilder. Both methods enable scarless construction and both allow for fast and economical ways to generate large and diverse combinatorial libraries.
For the assembly methods discussed here, the timeline for construction, validation, expression, purification, and analysis takes about seven days, and this is using traditional cell-based expression methods. In terms of scope, the average researcher using these methods could construct an assay of about 1,000 proteins every two work weeks. By simply switching to a cell-free expression method like NEBExpress, our E. coli lysate-based product, or PURExpress, our reconstituted transcription translation system, the timeline for construction to analysis is cut in half to about three days total. Furthermore, since these cell-free systems are easily dispensable by most automated liquid handlers, there's no limit to throughput. So now that same average researcher that adopts these cell-free methods can handle tens of thousands of proteins each week.
Before we go any further, I'd just like to define high-throughput. High-throughput means different things to different people and ultimately it just means more. If you're used to working with tubes, then plates are high-throughput. If you're accustomed to plates, then maybe you think high-throughput is droplet-based methods. Here, I will be focusing on the 96 to 384 well range.
The variety of equipment used for high-throughput automation is tremendous, but essentially boils down to four essential tasks: bulk fill, bulk transfer replication, cherry-picking or complex mixing, and signal detection. We accomplished these tasks with electronic multichannel pipettes, the Integra VIAFLO, the Echo 525 Acoustic Liquid Handler, and the Biotek Neo2 Plate Reader respectively. We find this suite of instruments sufficiently supports high-throughput interrogations while also enabling researchers to rapidly customize approaches for the diverse and ever-changing needs.
The star of the show here is Echo 525 Acoustic Liquid Handler. The Echo 525 uses acoustic waves to propagate the transfer of 25 nanoliter droplets from a source plate to a destination plate. These transfers are scripted, rapid, and robust, meaning it can support a variety of complex solutions. In addition to complex mixing, its ability to precisely transfer small volumes allows the user to miniaturize a diverse range of reactions including PCRs, ligations, transformations, and cell-free protein synthesis solution. This feature becomes essential as we scale up to reduce costs and overall waste, and here we'll discuss how to use the Echo to generate large complex arrays of design constructs.
By adopting an automated and miniaturized platform for high-throughput construction and expression, researchers can perform more reactions in less time and do so more accurately.
All right, so let's get started.
I will begin by discussing NEBridge Golden Gate Assembly. In this example, we will assemble six synthesized fragments into a closed circular plasmid. I would like to note that ORF construction or the assembly of a gene of interest and insertion into a vector backbone utilizes the same exact approach just with different designs and fragment length. Both approaches use a restriction ligation method utilizing Type IIS restriction enzymes.
Unlike traditional restriction cloning with Type IIP restriction enzymes, Type IIS restriction enzymes cut outside of the recognition site leaving a user-defined four base pair overhang, thus enabling scarless cloning of many fragments at once.
Here is an example of six fragments labeled A through F, each with complementary ends that would allow for seamless assembly into a closed circular plasmid at the six complementary junction sites. The primary limitation to this method is understanding the compatibility and/or crosstalk between different overhangs at these junctions to enable the most efficient ligation.
To better understand this limitation, scientists at NEB ligated SMRT-bell fragments with degenerate four base pair overhangs and performed deep sequencing. The heat map on the right illustrates the results. The strongest ligation efficiency is presented along the diagonal suggesting that fragments with complementary Watson-Crick base pairing ligated the most efficiently. Hotspots outside of this diagonal signify mismatched ligations or non-complementary overhang successfully ligated.
From this data, we were able to compute new rules for Golden Gate Assembly, which we refer to as Data-Optimized Assembly Design, or DAD. If you listen to DAD, one can now achieve much higher efficiency when working with a larger number of fragments, and this really opens up the possibilities accessible to researchers.
This work has been published extensively in numerous articles over the last two years and I encourage you to check them out if you would like to learn more. Of particular note is a 2022 publication from Pryor et al. that successfully performed a 52-piece assembly to reconstruct the T7 genome.
Additionally, we now offer multiple web tools. Ligase Fidelity Viewer helps viewers score their existing designs, GetSet defines sets of overhangs for use in a modular assembly strategy, and SplitSet splits the gene into multiple fragments that optimize junction sites. Here we'll use GetSet to define six optimized junctions for our six-piece assembly.
Prompting GetSet with our desired restriction enzyme incubation condition, the number of desired overhangs gave us CGGA, CAGC, ACGA, and so on. This set is optimized to have 100% fidelity.
Plugging the optimized junctions into our design looks like this, such that Fragment A has Overhang 1 CGGA on the left side, and the reverse compliment of Overhang 2 GCTG on the right side. What's great about this design approach is that we can now apply these same junctions to a variety of fragments of each type.
In this way, a single design can be used to construct and interrogate a multitude of plasma designs with a variety of promoters, 5'UTRs, and terminal tags, genes of interest, antibiotic resistance markers, and origins, all with high efficiency and high fidelity. With five variations of each piece in a six-piece assembly, the full factorial library consists of more than 15,000 unique designs. As you can imagine, the technical aspect of assembly of these constructs now becomes the difficult part, so let's walk through our approach.
Once the fragments are designed and ordered, we typically amplify and purify our fragments of interest. This results in highly pure linear fragments that define concentrations. Using fragments straight from the manufacturer can be successful, but we find the amplification and purification steps greatly increase the efficiency. We further note that the highest efficiency can be achieved by using pre-cloned donor vectors.
Now that we have the pieces, we use the complex and scripted mix and feature of the Echo 525 to combine the pieces accordingly such that each well of the resulting plate has all the fragments required to assemble a unique plasma design. We then bulk-fill our one-pot NEBridge Golden Gate Assembly mix on top of the combined pieces, thus generating our closed circular products. Notably, we can take advantage of the Echo to miniaturize the NEBridge reaction 50 times the standard view of volume.
Now that we have the plasmas constructed, we need to transform them into a cloning string like NEB 10-beta. Conveniently, we sell these cells in large formats specifically for applications like these high-throughput transformations.
Once transformed, we use our bulk replicator to make serial dilutions and perform spot plating. Shown here are two different dilutions of the same 96-well transformation where each spot shows multiple colonies for each unique plasma design.
Now that we have propagated our plasmids, the next thing we do is pick colonies, perform sequence validation, cherry-pick the confirmed clones, and proceed to protein purification. A traditional protein expression using columns generates a high yield and high purity, but they tend to be rather slow and tedious and they're not really compatible for high-throughput.
High-throughput protein purification at Biolabs involves retransformation of validated constructs into expression strains, expression in deep-well plates, and a one-pot lysis and retrieval protocol on the KingFisher Flex, which is essentially an automated magnetic pin tools. None of this would be possible without a number of additional products that NEB offers, including special expression strains that are amenable to auto induction and magnetic nickel beads that are compatible with our cell lysis solution that enables purification of an uncleared lysate.
Shown here is a Coomassie stain SDS-PAGE gel of a high-throughput purification using titrated amounts of magnetic nickel beads. From this, you can get an idea about the achievable purity and yields from our automated high-throughput protein purification of 1 mL cultures.
The only problem with this procedure is that it's not easily scalable. So While it's great for hundreds of proteins, for the modular NEBridge design I presented earlier that comprise more than 15,000 designs, we need a better way to handle the throughput. To increase the throughput of our expression and also decrease the overall time for each project, we have turned the cell-free protein synthesis. Before I get into cell-free protein synthesis, I'd like to present our second approach for generating mutagenic libraries for screening.
In contrast to assembling synthesized fragments, we can amplify an existing plasmid with mutagenic primers and then assemble the generated fragments with homologous ends using NEBuilder.
In order to simplify the explanation of multi-site mutagenesis, I will first describe site-directed mutagenesis using a single pair of primers that can be achieved with our Q5 Site-Directed Mutagenesis kit. Site-directed mutagenesis uses two adjacent primers with one containing the mutation of interest. In this case, that's the forward primer on the top. These primers are used to amplify the sample template and the PCR product is then treated with a kinase, ligase, DpnI mix, or KLD, to perform a blunt-end ligation turning sample to simple in a couple of hours.
Primer design is straightforward, but for a single project where the hundreds of primer sets are needed to be generated, an automated design tool is required. For many years, we've offered an interactive primer design tool in the form of NEBase Changer where users can upload their sequence of interest and input their desired change to automatically generate primers for insertions, deletions, and mutations. We now offer a batch tool that allows users to input a list of multiple mutations for the same template, ultimately generating a table of automatically designed primers. The output for the Batch Mode looks something like this where each row specifies an individual mutation and contains both a forward primer containing the mutation and an adjacent reverse primer. For Q5 site-directed mutagenesis using KLD, we ordered these primers as dual sets with both primers in the same tube.
The advantage of ordering these sets together is that you can simply do a bulk transfer from the primer plate to a new plate and bulk-fill on top of that with our Q5 Master Mix containing the template of interest. It takes a minute to set up and an hour or two to run, and when it's done, we often do some sort of quality control checks like this gel here. As you can see, it's a robust and easy process.
With our PC products in hand, we go straight into our KLD reaction with no cleanup required. The standard volume for a KLD reaction is 10 microliters, but we've successfully miniaturized down to half a microliter, and I believe you can go much further. After a brief five-minute incubation at room temperature, we have achieved ligation and generated our closed circular plasmids. Next, we go straight to high-throughput transformation and plating, as shown previously for NEBridge. Same downstream protocol previously discussed for NEBridge is also used here, and as before, there's a significant time and throughput advantage to employing cell-free protein synthesis over traditional expression and purification strategies.
After completing our query into the single mutants, let's say we identify 20 or so that had the most significant positive effect. Our next step is to understand the potential cooperativity of these mutations and hopefully identify a multi-mutant with further enhanced activity. And here's where multi-site mutagenesis comes into play. In this example, we've decided to take these 20 hits and interrogate them using a large number of mutants, rated here in columns each containing five mutations. How we're going to do this as using mutagenic primers to amplify a known template. After PCR, we now have multiple fragments, each containing homologous ends to their respective counterparts that enable full plasma to be assembled via NEBuilder.
The forward primer used for each individual mutation is identical to that previously described for Q5 site-directed mutagenesis, but the reverse primer is different. Instead of it being adjacent, it is now the reverse compliment of the forward primer. Amplification with these primers together would produce a single fragment with homologous ends that could self-ligate to form of single mutation, and this would be an alternative Q5 site-direct mutagenesis. However, the beauty here is that multiple sites can be linked together. For a second mutation shown here in orange, the primer design is the same as the first; perfectly complimentary primers, both including the mutation of interest.
By amplifying the template with the forward primer of set one and the reverse primer of set two, we create a small fragment that has two mutations, one on each end, and homologous ends that will ligate with the amplification product of set one's reverse and set two's forward primer. In this way, we can link upwards of five or six mutations together just by bearing which primers they use together to generate which mutagenic fragments.
For primer design, we can use the same batch option for any base changer with two major differences. First, we need to avoid the adjacent non-mutation primer and instead reverse compliment the forward primer containing the mutation. Secondly, we need to order these primer sets in separate wells so that we can use them independently of each other. Again, these two primers can be used together for site-directed mutagenesis, but for multi-site mutagenesis, they need to be used separately.
Going back to our example from before and trying to gain insight into higher order gain-of-function mutants, it's important to note that 20 primer sets that we've ordered separately, so 40 primers total, can make more than 10,000 unique combinations using the described method of mixing and batching the forward and reverse primers for proximal mutations. It's an extremely cost-efficient way at probing mutational diversity, and as you will see, the technical process is rather straightforward.
Taking the first multi-mutant as an example, we first rank the mutations according to residue and identify the mutational combinations of each fragment. We then identify the primers needed to construct these fragments and their well positions, and we then iterate through each multi-mutant making sure to keep track of which fragments are shared between each mutant so as not to duplicate our efforts. As before, we amplify using our Robust Q5 Mix, and this time using the complex mixing feature of the Echo to distribute the individual primers appropriately.
We then must combine the appropriate fragments for each assembly also using the complex mixing feature of the Echo. Now each well should have the necessary fragments for NEBuilder to work its magic. The Master Mix of all NEBuilder components can then be layered on top of our combined fragments. Shown here in gray is the standard reaction volumes, but we have tested and successfully achieved an 80-fold miniaturization that both cuts cost and waste.
One thing I'd like to highlight is that even under this significant miniaturization, which reduces the cost per reaction down to a dime, we still achieve great efficiency and have plenty of colonies to validate for downstream expression.
Another technical point to take home is that minor adjustments in input concentration or relative volume can have significant effects on your transformation efficiency. Shown here is an Echo dilution plating of NEBuilder reactions. There are five different reactions with varying insert or vector ratio plated by row. In the highlighted data, a twofold increase in insert concentration resulted in tenfold increase in transformation efficiency. This also highlights another feature of the Echo and that is it's capable of treating cells and offers an alternative method for high-throughput plating.
For further information on NEBuilder, please visit our website and also lab sites material now available via Beckman Coulter. The top-left node is a particular interest as a detailed multi-site mutagenesis with NEBuilder.
As we've alluded to throughout the presentation, high-throughput construction methods only push the bottleneck to expression and purification and don't solve the whole problem. In order to take full advantage of the modular and combinatorial construction methods detailed thus far, we need to employ cell-free protein synthesis to meet throughput demand and accelerate our project timeline.
Cell-free protein synthesis systems supply in vitro mixtures that contain all the necessary components to support transcription and translation, including all the necessary enzymes, energy sources, and substrates. The user simply provides a template and goes from DNA to protein in as little as two hours. NEB has two cell-free protein synthesis systems available. NEBExpress, that is our E. coli S30 lysate-based product, and PURExpress, which is a reconstituted system with purified components. Both utilize T7 RNA polymerase for transcription and can accept both circular or linear DNA or RNA.
Integrating cell-free expression into high-throughput workflows often suffers from resource limitation. By miniaturizing our cell-free systems, we can increase the accessible throughput, thus removing this barrier. As we began to systematically miniaturize these systems, we were interested in determining what DNA input types work best, what if any volume constraints exist, and how did these factors affect reproducibility?
We performed miniaturization with the Echo 525, and since cell-free systems employ complex solutions, our first question was how well do our components shoot on this device and are the reactions assembled by the Echo as reproducible as those assembled by pipette? Now, all the assays from here on out employ the same construct whereby expression of a fast-maturing YFP protein is under control of the T7 promoter. We compare the two devices at the lowest reaction volume we could reliably pipette and found that the two were both similar and reproducible with CVs below 10%, suggesting that both NEBExpress and PURExpress components were shot accurately and precisely by the Echo.
Having confidence in the method, we next sought to determine the linear range of circular DNA as input for both NEBExpress and PURExpress. Both NEBExpress and PURExpress responses were highly sensitive to circular plasma DNA. The maximum yield of NEBExpress plateaued at 5 nM input, while PURExpress became saturated at 2 nM. We did the same for linear DNA in the form of a purified PCR product and observed almost identical results for PURExpress indicating that equimolar concentrations of closed circle or linear fragments performed similarly in the purified system. However, for NEBExpress, a threefold increase in the amount of linear DNA was required to maximize yield, likely owing to the presence of exonucleases in the lysate-based product.
Now that we know that linear ranges for DNA input concentration, we can compare the response from different reaction volumes and how they affect total protein yield. We tested three volumes, 2.5 microliters, 1 microliter, and 0.5 microliters. You can see here on the left that total yield scales with volume and that reproducibility is largely unaffected by miniaturization. That is to say, expression in 500-nanoliter reactions is reliable and reproducible. If we focus on the highlighted data and plot yield according to volume that's seen on the right, you can see that yield appears linearly dependent on volume such that a 0.5 microliter reaction produces half as much protein as a 1 microliter reaction.
While this dataset is only for any NEBExpress, the same trends were observed for PURExpress and ultimately these 500 nanoliter reactions represent a 50-fold, 25-fold volume reduction as well as similar increase in accessible throughput. It's worth mentioning that I've successfully miniaturized these reactions beyond the limits presented here so that these are even conservative reports.
I hope you can now appreciate how one can efficiently generate large modular or combinatorial libraries from this couple dozen synthesized fragments or mutagenic primer sets, and then translate them in vitro in half the time with no bottleneck save resources, which we have mitigated with miniaturized reactions. For further information on the miniaturization of our cell-free protein synthesis systems, I encourage you to check out our recent App Note entitled, "Scaling Down to Scale Up: Miniaturizing Cell-Free Protein Synthesis Reactions on the Echo 525 Acoustic Liquid Handler."
As an example of this high-throughput construction and expression protocol, I took a classic test case from Howard Salis's lab in which they varied the spacing region between the ribosome binding site and the start codon of the fluorescent protein. If the spacing is insufficient such that the ribosome can't see the start codon, it doesn't initiate, and if it's too far away, initiation is inefficient, so there's this Goldilock zone that promotes high-efficiency translation.
I copied this experiment generating constructs using our high-throughput construction methods, expressed the validated constructs in NEBExpress well above the saturating input concentration, and measured the fluorescence of triplicates all within three days of receiving the mutagenic primers, and here is the result. You can see that a similar trend is observed with the Goldilocks region centered around four to five base pairs. Strikingly, the reproducibility of these technical replicates was incredible. If we overlap the trend of the controlled data, you can see the tube overlap extremely well. This result not only validated our high-throughput construction and in vitro expression workflows, but also illustrate that NEBExpress translation, at least in terms of RBS spacing, mimics that of the cellular environment.
Interestingly, when we take the same exact clones and express them in PURExpress, we see a vastly different trend. As with the control sets, PURExpress yield suffers from a short RBS spacer, but a longer spacer fails to significantly affect the overall yield. This suggests that translation in PURExpress is not covered by the same principles as that in cells or NEBExpress, and nor should it considering its purified nature. This finding highlights the importance of picking the right tool for the job. If you want to mimic cellular expression, you may want to choose NEBExpress, but if you want unadulterated maximum expression irrespective of optimal construct design, you may want PURExpress.
As we conclude this seminar, I'd like to present the ultimate high-throughput construction and expression workflow that can be achieved in a single day. This protocol relies on the fact that all the cloning methods we've discussed thus far have included ligases and resulted in closed circular double-stranded DNA that is resistant to exonucleases. Instead of transforming our ligation products, we remove the unligated material and remaining junk with the next nuclease cocktail and reamplify the expression constructs using standard PCR. This product can then be passed directly into cell-free protein synthesis or be purified and then pass into synthesis. Since this method does not rely on clonal variants, but rather the amplification of a population of ligation products, high-accuracy cloning is required. We further note that other amplification methods like Rolling Circle Amplification, or RCA, can also be used to propagate your desired clone.
We repeated the same experiment described previously for interrogating the effect of spacing between the ribosome binding site and the start codon, but this time, all in vitro with no messy cells to deal with and all in one day. The resulting data shown here in black look very similar to the previous dataset I showed in light blue that used purified and validated plasma DNA. This approach is currently being optimized and we look forward to presenting our amazing results that have come from this workflow in the near future.
To summarize, I've presented high-throughput methods for modular plasmid and/or gene construction with NEBridge Golden Gate Assembly, site-directed mutagenesis with Q5 and KLD, multi-site mutagenesis with NEBuilder. I've also highlighted how to automate primer designs using our available web tools and how to efficiently miniaturize these reactions. After assembling your contract of interest, I've shown you how to achieve high-throughput protein purification utilizing our large-format competent cells, compatible cell lysis solution, and magnetic affinity beads. Lastly, I introduced our cell-free protein synthesis systems, NEBExpress and PURExpress, and showed how integrating these systems into your workflows increases throughput capabilities, efficiency, and reproducibility.
I hope you enjoyed today's seminar and on behalf of our company, I wish you the best of luck in your future experiments. Should you have any questions, we'll be running a live Q and A session after this, or additionally, please feel free to email me directly or call or text the support line. They're always here to help. Thank you.
Host:
Thanks, Jack. We also have a couple of our scientists here to help answer questions today. We're joined by Paula Magnelli, one of our Development Scientists, and Brad Landgraf, Senior Production Scientist, but we're going to kick it off with we have a few questions for Jack. I'm going to pass it over to you.
Jackson Buss:
Hello, everyone. Thanks for asking these questions. So the first one I have here is, "Does transformation method influence NEBuilder assembly efficiency?" And then apparently it doesn't work with some of our competitor competent cells. And I would just like note, yes, we vet all of our construction methods with our own competent cells, our 5-alpha and our 10-beta, and we see about 80% efficiency in terms of NEBuilder ligation per insert, and that's pretty well-documented on our webpage for NEBuilder. So please visit that for more information.
Here's another question here, "Is there NEB protocol available for mutagenesis?" And yes, again, please visit our website. There's Q5 site-directed mutagenesis. There's protocol pages for that as well as NEBuilder. And then specifically for the NEBuilder multi-site mutagenesis, there's an App Note for that which I highlighted in these slides, so if you go back a few slides, you should be able to see that.
Lastly, here we have, "What length could you use the Q5 site-directed mutagenesis for? Is 15 kb plasmid okay?" And the answer to that is yes, we don't know the exact limit, but we've tested upwards of 20 kb for Q5, so it's dependent on the polymerase. And you might lose a little bit of efficiency because ligating bigger products is less efficient sometimes, but yeah, we've done 20 kb, so the answer to that is go for it.
I'm going to pass it on to Brad here.
Brad Landgraf:
Thanks, Jackson. Another question we have is, "What product would you suggest for bacterial extract-based in vitro transcription and translation?" And for that, we would recommend our NEBExpress Cell-free E. coli Protein Synthesis System. It's a very robust product based on and optimized for T7 RNA polymerase protein production. So that's the product that we would recommend for bacterial in vitro transcription and translation.
I'll pass it over to Paula.
Paula Magnelli:
Thank you, Brad. Good afternoon, everyone.
I have here another question which is asking about what are the yields of the target protein using both NEBExpress and PURExpress? With either one, the yield on average is around 0.5 to about 1 milligram per ml, and of course, this is target-dependent. There is a range, but those are, in general, the yields we see using a variety of different targets of different lengths and confirmation and origin. I'll give it back here to Jack.
Jackson Buss:
All right. Thanks, everyone. Yes, we've got a couple more questions here. One is, "Can you also perform multi-site mutagenesis with NEBridge Golden Gate instead of Builder?" And the answer is yes. It's been published before and we actually will have an App Note coming out, hopefully in the near future, maybe the fall, specifically detailing this protocol and that you would just include the mutations in the primers with overhangs either outside the mutagenic region.
Moving on, we have one here from... "Are there any disadvantages to using a cell-free expression system in the workflow described versus the more traditional transformation expression cell lysis method?" So if we're talking about resulting in lysates, I would say no, there aren't really any disadvantages. This is very fast and it's robust and consistent from sample to sample. However, cells obviously enable you to scale up for a single protein very, again, high purity, high yield. So if that's what you're looking for, yeah, that's the way to do it.
Okay. Here we have, "NEBridge Golden Gate Assembly was performed for both the plasmid or ORF in the presentation. Could you use Builder instead for the..." Okay, so can you use Builder to assemble synthesize fragments? And the answer is yes, you certainly can. It's slightly lower efficiency, slightly more errors, and we put a limit on our Builder for about five to six fragments where Golden Gate Assembly can do, again, more than 52 and that we're still building out that a bit. I think the theoretical limit is like a 100. So yes, if you're just doing five or six fragments, Builder is fine, but Golden Gate will be more efficient.
Okay. Lastly... Oh no, we've got a couple more. I'll answer this and pass it on to Paula.
"You've highlighted and focused on the Echo 525 for small volume transfer and complex mixing. Are there other devices that are compatible and recommended?" So yes, we really like to Echo 525 because it's extremely precise and we like that tip list. But yeah, for small volume, we've tested the Formulatrix MANTIS and we know customers have used the mosquito LV. And then complex mixing, we've tested Opentrons' OT-2, and again, customers have used the Integra ASSIST, but there's plenty of other ones. Basically, we don't know of any automated liquid handlers that our reagents are not compatible with. Again, this isn't a comprehensive list, but yeah, let us know if you have problems, we'd work on it. But yeah, as far as we know, it's compatible with everything. Okay.
Paula Magnelli:
Thanks, Jackson. I have here a very good question, whether we can use the Cell-Free System for multi-unit proteins such as antibodies. Antibodies are usually better expressed in eukaryotic systems, although it is known that they can be expressed in bacterial systems, and in some cases, in cell fleets. However, the system, it is compatible with expression of, in general, multi-unit proteins because it's very amenable to express in equimolar amounts different polypeptides from independent classmates or constructs in a much, much easier way than what you will have to do in vivo.
So for that purpose for expressing multi-unit proteins, it's quite compatible with cell-free protein expression. Having said that, the case of antibodies because of their folding and post-translational modifications and the sulfate formation, it can be challenging in general to express in bacterial systems, although we know that that is possible, but it requires quite a level of optimization to achieve in a bacterial system.
Jackson Buss:
All right, yep. They keep coming in, so keep sending them. All right.
So I got a question here. "Have you done any testing with the Echo 555 and are there any restrictions related to the Echo compared to the Echo 525?" So short answer is no, I have not. I do know that. So I think there's three Echos, the 650 also. I know that the 650 doesn't work. That's DMSO bases, like small molecules. And so I'm seeing here the Echo 555, that it can do DNA and if it can shoot DNA, then it should be compatible. I don't know the... It's 2.5 nanoliters, so yeah, it sounds like the 555 is the new version of the 650, but for DNA. So yeah, it sounds good.
Okay. "How are you completing sequencing in the one-day workflow? What method are you using? We use a lot of amplicon ONT sequencing, and we have our own internal analysis pipeline. What automation or high-throughput platform did you use for E. coli transformation?" So first of all, the Echo can plate. So plating is the issue here, it's just volume transfer and then plating. So yeah, we use the Echo. We also use the VIAFLOW. We can use multi-channel pipettes. Yeah, nothing super fancy.
And then lastly, "Thanks for the seminar. I'm starting an AI project for the design of proteins by deep learning. Do you think that high-throughput cloning kits would be beneficial for cloning many sequences compared to traditional?" Of course, this is exactly what it's suited for. Either order them or have them synthesized or make them, and then yeah, throw them in here. As long as you have a quick assay or just seeing them expressed as the answer, then yeah, this is exactly the use case we're thinking of.
Okay, a couple more for Paula.
Paula Magnelli:
Hi, thank you. I have one here to clarify about expressing multiple targets at the same time like YFP and GFP, and indeed if the targets are in the same context, meaning in the same classmate with the same UTR and regulatory sequences upstream, so the only difference is the open building frame itself, coexpressing shouldn't be biased. It's more dependent on the regulatory elements. Therefore, if you are coexpressing two different proteins at the same time, they are roughly, in our experience, expressed at the same equimolar levels.
And related to that also whether the expression efficiency changes with increasing the size of the target, and we have seen that that is not the case, that the proteins, either small or large, are expressed at the weak, both PURE and NEBExpress at high levels without truncation in the case of large proteins. However, disulfide bonds or folding can... Difficult targets might have a propensity to meet fault, but that is, again, on a case by case basis.
Host:
Wonderful. Thank you. We are coming up on time, so if we didn't have a chance to get to your question today, we'll be sure to follow up with you offline. If you do have any other questions that come up for the webinar, Jack's email is up here on the screen, but also you can feel free to email webinars@neb.com. This presentation will also be available on-demand after today's presentation and I hope you all have a great day. Let us know if you have any questions. Thanks.
Related Videos
-
Introduction to NEBUILDER HIFI DNA ASSEMBLY® -
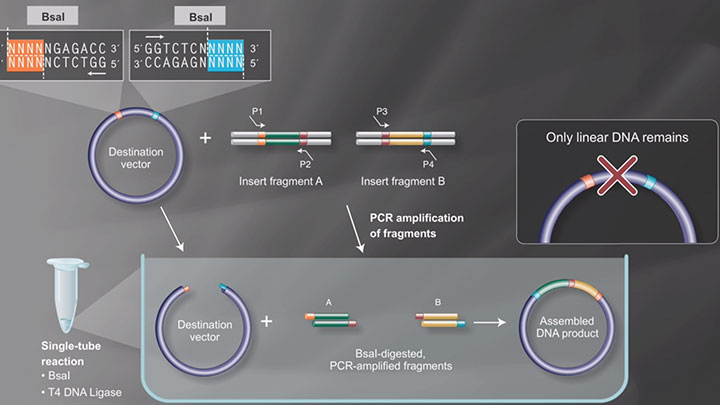
Golden Gate Assembly Workflow

