A Breakthrough Method of Small RNA Sample Preparation for Next Gen Sequencing
Script
Daniela Munafo:
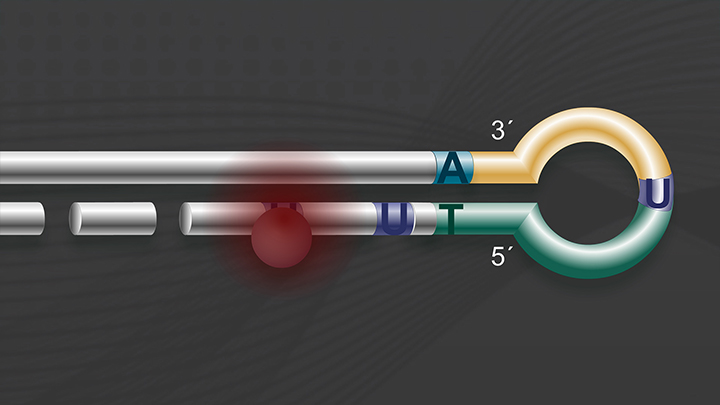
Most methods for small RNA library constructions are based on sequential ligations of adapters that leads to adapter dimer formation.
Daniela Munafo:
Adapter dimers can be a real problem because it strongly contaminates the library, it reduces the library yield, and increases background.
Brett Robb:
So there are a number of strategies to get around the adapter dimer problem in library construction. Traditionally the first approach would be to gel purify in between ligation steps. This has the problem of running multiple gels, and also takes more time and reduce the complexity in your final libraries.
Brett Robb:
The second approach is to reduce the adapter concentration in the ligation reactions. This again can result in reduced complexity of your library sent for sequencing.
Brett Robb:
The third approach would be to use enzymatic steps to inactivate the adapters in the ligation steps. So when you get to the final step of library purification, often the major species on your gel is the adapter dimer product.
Brett Robb:
One of the consequences of purifying the adapter dimers is that you reduce the complexity in your sequencing library and essentially end up with junk data.
Daniela Munafo:
So we were, at the meeting, trying to solve this problem when Larry says, "Hey, what if we switch the order of the steps?" For the first time we solved this problem without having to introduce any more enzymatic steps, neither more gel purifications. By transforming single strand adapters into double strand forms, we improved performance and increased the sensitivity of the small RNA library construction.
Related Videos
-
NEBNext® Ultra™ II Directional RNA Workflow -

Optimization of NGS Library Preparation: Low Inputs and Fast, Streamlined Workflows

