Characterization of mRNA modifying pseudouridine synthases
Covalent chemical modifications are one of the most conserved feature of RNA molecules. Total number of RNA modifications in all three domains of life currently exceeds 250. Among those Pseudouridine (9U or Y) is the most prevalent RNA modification occurring in both coding and non-coding RNA. This RNA modification is installed by a family of protein called pseudouridine synthases. However, RNA preference of these enzymes is not clear. It’s not known what the structural requirements for the substrate are. Kinetics parameters of the modification and cofactors of the reaction have not been determined. To answer these questions, we use biochemical, structural biology and mass spectrometry methods to further characterize the candidate enzymes.
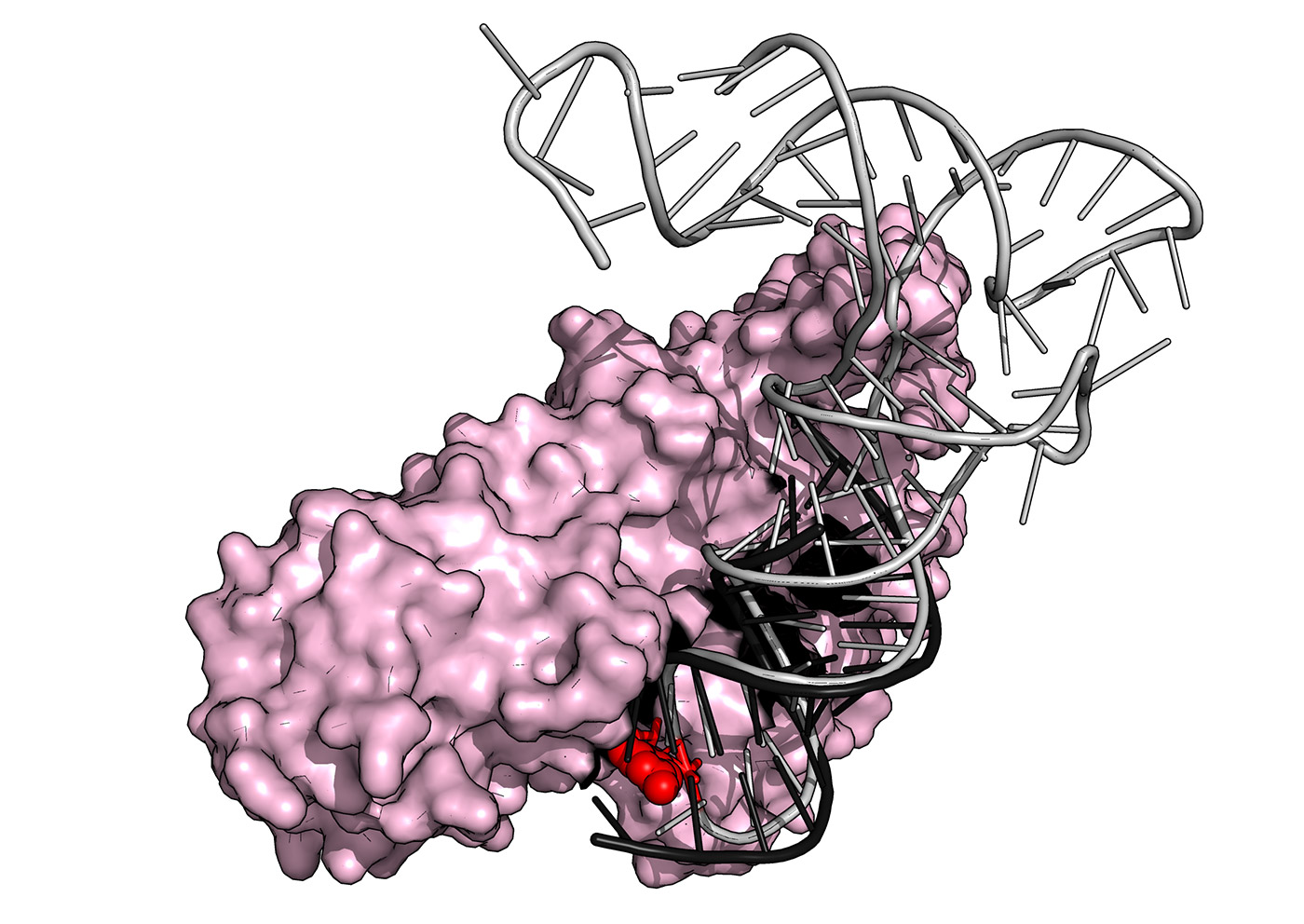
Superposition of the crystal structure of yeast PUS1 (pink) bound to an RNA substrate (black) with a human tRNA (grey) known to be modified by PUS1 within its anticodon loop (red sphere). The structure was solved by our group in collaboration with The Barry Stoddard Lab at the Fred Hutchinson Cancer Research Center.

