Eukaryotic Transcription Start Sites (TSS)
Building on Cappable-seq, we have developed ReCappable-seq to capture, in addition to non-Pol-II transcripts, 7-methyl G-capped transcripts derived from Pol-II RNA polymerase. To achieve this, we employ the yeast scavenger decapping enzyme (yDcpS) to convert capped RNA into di-phosphorylated RNA that can be “re-capped” by VCE. ReCappable-seq identifies TSS for RNA transcripts derived from all RNA polymerases.
In collaboration with other labs at NEB and the Akeson lab at UCSC we developed a Nanopore sequencing method called NRCeq. We identified individual full-length human RNA isoform scaffolds among millions of nanopore poly(A)-selected RNA reads. To achieve this we identify RNA strands bearing 5′ m7G caps, by exchanging the biological cap for a modified cap attached to a 45-nucleotide oligomer. This oligomer adaptation method improves 5′ end sequencing and ensures correct identification of the 5′ m7G capped ends.
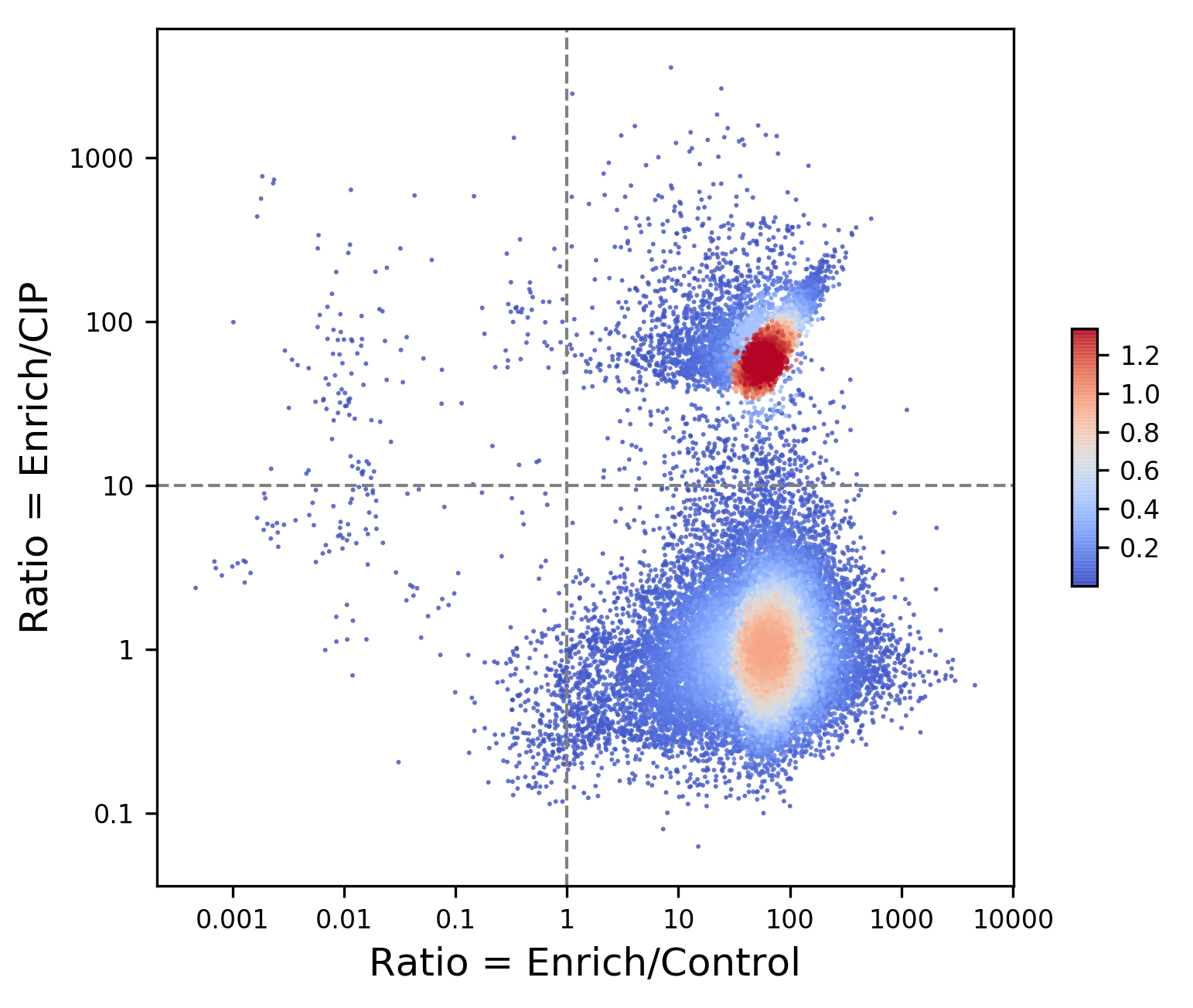
Global distribution of Transcription Start Sites (TSS) from Jurkat cells analyzed by ReCappable-seq. The two groups represent Pol-II TSS (lower cluster) and non-Pol-II TSS (upper cluster). Each dot represents one genomic position. Streptavidin enrichment ratio is plotted along the x-axis and CIP sensitivity ratio along the y-axis.

