Library Preparation Using NEBNext® Multiplex Small RNA Library Prep Set for SOLiD™ (Set 1) (E7450)
 This is
a point where you can safely stop the protocol and store the samples at -20°C
for up to 72 hours.
This is
a point where you can safely stop the protocol and store the samples at -20°C
for up to 72 hours. This caution sign
signifies a step in the protocol that has two paths leading to the same end
point but is dependent on a user variable, like the amount of input DNA.
This caution sign
signifies a step in the protocol that has two paths leading to the same end
point but is dependent on a user variable, like the amount of input DNA.- (green cap) 3´ SR Adapter 3
- (pink cap) SR RT Primer 3
- (yellow cap) 5´ SR Adapter 3
Ligation of 3´ and 5´ Adaptors (~1 hour and 15 minutes to the first safe stop point)
- Mix the following components in sterile nuclease-free strip tubes:
Input RNA 1–6 μl
NEBNext 3´ SR Adaptor 3 1 μl
1 μl
Nuclease-free Water variable
------------------------------------------------------------------
Total volume 7 μl
- Incubate in a preheated thermal cycler at 70°C for 2 minutes and transfer
tube to ice.
- Add the following components:
3´ Ligation Reaction Buffer 10 μl
3´ Ligation Enzyme Mix 3 μl
------------------------------------------------------------------
Total volume 20 μl
- Incubate at 25°C for 1 hour in a thermal cycler.
 The sample can be safely
stored at -20°C for 72 hours at this point. ~1 hours and 15 minutes to the next
safe stop point.
The sample can be safely
stored at -20°C for 72 hours at this point. ~1 hours and 15 minutes to the next
safe stop point.
- Add the following components to the ligation mixture from step 4 and mix
well:
Nuclease-free Water 4.5 μl
SR RT Primer 3 1
μl
1
μl
------------------------------------------------------------------
Total volume 25.5 μl
- Heat samples at 75°C for 3 minutes then ramp to 4°C at a rate of 0.3°C/sec
and transfer to ice.
- Resuspend the 5´ SR adaptor 3 in 35 μl Nuclease-free Water. Heat the
resuspended adaptor at 70°C for 2 minutes and transfer to ice (only heat denature once).
- Add the following components to the ligation mixture from step 7 and mix
well:
5´ SR Adaptor 3 (from Step 7) 1 μl
1 μl
5´ Ligation Reaction Buffer 1 μl
5´ Ligation Enzyme Mix 2.5 μl
------------------------------------------------------------------
Total volume 30 μl - Incubate at 25°C for 1 hour in a thermal cycler.
 The sample can be safely
stored at -20°C for 72 hours at this point. ~1 hour and 30 minutes to the next
safe stop point.
The sample can be safely
stored at -20°C for 72 hours at this point. ~1 hour and 30 minutes to the next
safe stop point.
- Mix the following components in sterile nuclease-free strip tubes:
3´→5´ Ligated RNA from step 9 14 μl
NEBNext First Strand Synthesis Reaction Buffer 4 μl
Murine RNase Inhibitor 1 μl
------------------------------------------------------------------
Total volume 19 μl
- Heat the sample at 42°C for 2 minutes and then add the following component.
Preheated RT buffer and sample mix 19 μl
ProtoScript II Reverse Transcriptase 1 μl
------------------------------------------------------------------
Total volume 20 μl - Incubate mixture for 42°C for 1 hour and then at 70°C for 15 minutes.
 The sample can be
safely stored at -20°C for 72 hours at this point.
The sample can be
safely stored at -20°C for 72 hours at this point.
- Mix the following components in sterile strip tubes:
RT reaction mixture (from step 12) 20 μl
OneTaq Hot Start 2X Master Mix 25 μl
Index Primer (X)* 2.5 μl
SR Primer R3 2.5 μl
Total volume 50 μl
------------------------------------------------------------------
* Note: The kit contains 16 index primers, each with a unique index. For each reaction, only one Index Primer is used.
- PCR cycling conditions:
CYCLE STEP TEMP TIME CYCLES Initial Denaturation 94°C 30 sec 1 Denaturation
Annealing
Extension94°C
60°C
65°C10 sec
30 sec
15 sec12-15 Final Extension 65°C 5 min 1 Hold 4°C ∞
Size Selection of Amplified cDNA Library
- Add 10 μl of Gel Loading Dye, Blue (6X) to each amplified cDNA construct (60
μl total volume).
- Load 5 μl of Quick-Load pBR322 DNA-MspI Digest in one well on a 6% PAGE gel.
- Load each amplified cDNA construct with loading dye by splitting into two wells (30 μl each) on the 6% PAGE gel.
- Run the gel at 120 V until the front of the dye reaches the bottom of the
gel (~60 minutes). Do not let the dye exit the gel.
- Remove the gel from the apparatus and stain with SYBR Gold nucleic acid gel
stain in a clean container for 10 minutes on orbital shaker and view the gel on
a UV transiluminator.
- Cut the bands corresponding to ~110–119 bp, which correspond to
adaptor-ligated constructs derived from the 21 and 30 nucleotide RNA fragments,
respectively. DO NOT cut the 89 bp band out, as this is adaptor dimer (Figure 1).
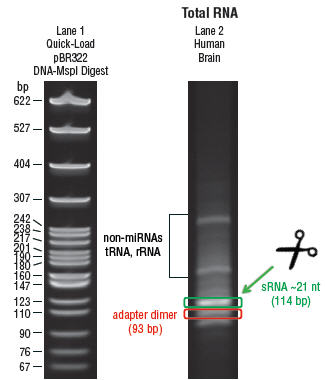
Figure 1: Small RNA Libraries
Small RNA library generated from 5 μg Human Brain Total RNA (Lane 2) and 5 μl of the Quick-Load pBR322 DNA-MspI Digest (Lane 1). The 114 nucleotide band in Lane 2 contains prepared miRNA library generated from ~21 nucleotide small RNA fragments. The 93 nucleotide band in Lane 2 contains adapter dimer
- Place the gel slice in a 1.5 ml tube and crush the gel slice with the
RNase-free Disposable Pellet Pestles and soak in 100 μl DNA Gel Elution Buffer
(1X).
- Rotate for 2–18 hours at room temperature.
- Transfer the eluate and the gel debris to the top of a gel filtration
column, and centrifuge the filter for 2 minutes at 14,000 rpm.
- Check the size (should be between 110-130 bp), purity and concentration on
an Agilent 2100 Bioanalyzer® (Agilent Technologies, Inc.) using a DNA high
sensitivity on a DNA 1000 CHiP. If the gel doesn't run properly for your sample
perform the following ethanol precipitation.
- Recover eluate, add 1 μl Linear Acrylamide, 25 μl 3M sodium acetate pH 5.2
and 750 μl of 100% ethanol and vortex well.
- Precipitate in a dry ice/methanol bath for at least 30 minutes then spin in
a microcentrifuge (>14.000 x g) for 30 minutes at 4°C.
- Remove the supernatant taking care not to disturb/remove the pellet and wash
the pellet with 80% ethanol by vortexing vigorously.
- Spin in a microcentrifuge (>14.000 x g) for 30 minutes at 4°C.
- Air dry pellet for up to 10 minutes at room temperature to remove residual
ethanol.
- Resuspend pellet in 10 μl TE Buffer. Perform the following quality control
analysis on your sample library to quantify the DNA concentration.
- Load 1 μl of the reconstituted construct on an Agilent 2100 Bioanalyzer
using a DNA High Sensitivity or an Agilent DNA-1000 chip (Figure 2).
- Check the size (should be ~110–119 bp), purity and concentration of the
sample. The final product should be a distinct band. If you see undesirable
peaks (bigger or smaller than your expected range sizes) perform a second round
of size selection.
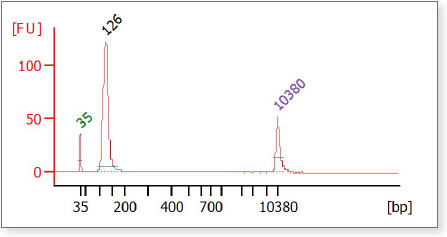
Figure 2: Agilent Bioanalyzer Trace of a final Human Brain miRNA Library showing a 126 nM peak peak between 114 and 123 bp.

