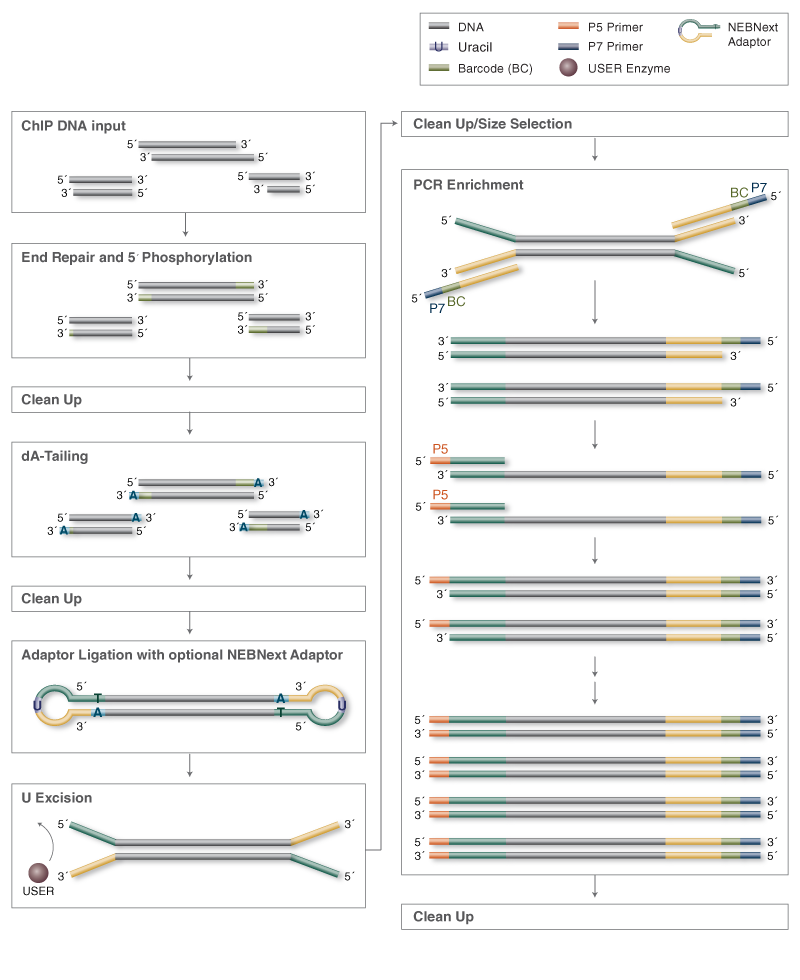
ChIP-seq DNA Library Preparation
Return to DNA Library Prep for IlluminaPreparation methods for Chomatin-immunoprecipitated (ChIP-seq) DNA libraries are similar to those for standard DNA. However, the amount of input DNA for ChIP-seq libraries is lower than for standard DNA library construction, and library preparation requires the use of optimized reagents and protocols. Choose the ChIP-seq Library Construction Workflow tab below to see specific workflows for the preparation of ChIP-seq libraries for both Illumina® and SOLiD sequencing. The isolated ChIP-seq DNA is treated to remove overhangs and add 5′ phosphates and 3′ hydroxyls. Libraries to be used with the SOLiD™ 4 platform can be directly ligated to adaptor after end repair and size selection. For Illumina® and Quick 454™ libraries, there is an addition of a dA-tail before ligation to an adaptor. Libraries are then amplified by PCR.
ChIP-seq Library Construction Workflow