Using large-scale RNA sequencing to probe immune response, including autoimmune disease and vaccination
Posted on Wednesday, October 13, 2021
By
Topic: What is Trending in Science
Immune repertoire is a term used to define the sum of an individual’s expressed T and B cell receptors (TCRs and BCRs, respectively). It is a dynamic state, encompassing the scope of an immune response. The TCR/BCR expression profile contains a wealth of information about the mysteries of immune responses in health and disease from both recent and past exposures. The information can be used to tease out immune cell involvement in autoimmune disease or to evaluate a host immune response to vaccination. Also, because different diseases (and even disease severity) can lead to a specific immune profile, it is clinically relevant due to potential utility in directing treatment choices and predicting treatment efficacy.
Gaining insight into autoimmunity with immune sequencing
Interesting research conducted in the lab of Associate Professor Kevin O’Connor from Yale School of Medicine is a great example of how multiple immune sequencing techniques can be leveraged to make huge leaps in understanding autoimmune disorders.Myasthenia gravis (MG) is one of the diseases investigated in the O’Connor lab. It is an autoimmune disorder that affects neuromuscular transmission and causes muscle weakness in the eyes, mouth, throat and limbs. Its pathological mechanism arises at the neuromuscular junction and is well researched and understood. Briefly, autoantibodies bind to the acetylcholine receptor (AchR) and interrupt normal signal transmission at the neuromuscular junction. Different types of MG are defined according to the type of autoantibody that binds to the various complexes that make up the AchR.
There were two questions the O’Connor lab specifically wanted to answer:
1. Why is thymectomy only partially successful in treating MG?
2. Why do MG patients treated with Rituximab relapse after a period of remission?
To answer these questions, scientists in the O’Connor lab employed multiple complementary immune sequencing techniques, resulting in a more complete picture of the immune response. To understand how these methods work together, we need to understand some basic immunology, as well as how these methods work.
Immune receptor diversity is discoverable in mRNA
TCRs and BCRs are expressed on the cell membrane, although B cells also secrete the immunoglobulin portion of the BCR as antibodies that can bind directly to antigens.An incredible level of immune receptor diversity of B and T cells is necessary to recognize an almost infinite number of antigens. This diversity is accomplished through somatic recombination that takes place at the DNA level in the heavy and light chains of BCRs, and the α and β chains of TCRs.
The V(D)J and constant (C) region of BCRs and TCRs look like this:
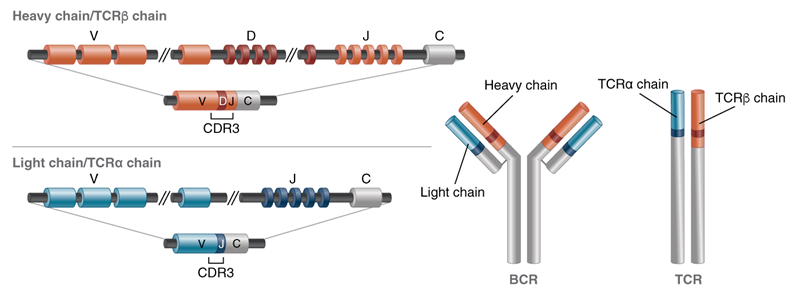
Figure 1: Simplified representation of the structure of an antibody or TCR showing the outcome of V(D)J recombination in mature lymphocytes.
Somatic recombination is accomplished through V (variable), D (diversity), J (joining) recombination; each cell selects a single V, D and J gene (and there are many of each). The recombination process also introduces non-germline-encoded nucleotides at the junctions, which further increases the diversity and specificity. These gene combinations are so random that it is unlikely that two cells will have the same sequence, so it essentially serves as a barcode. The constant region (C) is adjacent to the J region and also makes up a part of this unique fused gene signature.
Traditional methods used to detect TCRs and BCRs cannot capture the enormous sequence diversity of our immune cells – managing only a fraction of what we can now achieve with increased read-length and throughput of next-generation sequencing platforms.
Scientists at New England Biolabs® have taken immune repertoire sequencing to a new level with a workflow that enables bulk mRNA sequencing of the variable regions of BCR heavy and light chains and TCR α and β chains in humans or mice generating full-length UMI-tagged sequences.
What information can we gather from bulk, full-length sequencing of expressed immune receptors?
There are multiple aspects of bulk sequencing that improve the view on immune response.
Improved sequence accuracy - A bulk sequencing approach generates libraries from several million cells in one tube to maximize the sequence profiling of the immune repertoire. Each mRNA molecule is reverse transcribed into cDNA, which is tagged at the 3’ end with a unique molecular identifier (UMI), so each mRNA molecule present in the sample is sequenced. Multiple copies of an individual mRNA can be collapsed into a single consensus sequence, improving sequence accuracy.
Isotype characterization - Full-length sequences are generated to include both V(D)J region and constant (C) region. The ability to sequence the V(D)J region and also incorporate the C region is significant because, while the V(D)J region carries the most diversity, it doesn’t capture all of the variability; in B cells, the C region is where we find information that identifies isotypes (IgM, IgD, IgG, IgA and IgE) and more detailed categories of immune cells.
If a B cell is antigen-driven, immature immunoglobulin M (IgM) undergoes class-switching to antigen-experienced (or activated) IgD, IgG, IgA and IgE (all of which differ at the sequence level in the constant region). This is informative for translational research because isotypes can change throughout the course of an infection.
Access to full-length sequence information also allows for downstream antibody synthesis and functional characterization, which is not possible when sequencing only the V(D)J region.
Origin of clonal lines - Expansion of a single clone can be documented. Each level of the ‘clonal family tree’ is based on the previous clone, but it also contains sequence information about somatic mutations that have accumulated in the variable region. By aligning the sequence with the germline sequence, insight can be gained about the tissue origin of a particular clone providing a greater understanding of the immune response in disease states.
Single-cell and bulk immune cell RNA sequencing complement each other
Bulk sequencing generates BCR and TCR libraries with high throughput that allows a library of millions of cells to be prepared in one tube and sequenced with a relatively low-pass to screen the clonal diversity of the samples, whereas single-cell sequencing falls in the range of ~10,000 cells per library preparation and requires much higher sequencing coverage, which greatly increases the cost of library construction and sequencing.
On the other hand, single-cell sequencing enables a researcher to pair the heavy and light chain in B cells to identify a single B cell signature, or pair α and β chains in T cells to identify a single T cell signature. So, it gives a greater depth of information at a single-cell level.
So, how were these sequencing techniques used to answer the questions posed by the O’Connor lab?
1. Why is thymectomy only partially successful in treating MG?
In one MG subtype, there is thymus involvement, and thymectomy is used as a treatment but is only partially successful, and the O’Connor lab wanted a greater depth of understanding as to why.
In one study, they gathered full-length, bulk sequencing information from thymus tissue and patient blood that was longitudinally sampled pre- and post-thymectomy. They traced specific autoantibody-producing B cell clones to their tissue origin AND they bulk categorized immunoglobulin isotypes. When full-length B cell sequences were aligned in a clonal family tree analysis fashion, they discovered that some autoantibody B cell clones had expanded and were found in both the thymus and the blood. Additionally, they were able to deduce that B cells from the thymus were almost 100% IgG – so they had undergone class-switching from immature IgM upon antigen exposure; yet, there was a more even distribution of IgM, IgA and IgG in the blood. This suggests antigen exposure occurred in the thymus. So, the model that they were able to establish was that there were autoantibody-producing B cells present in the thymus that had clonally expanded and were also present in the blood. Following removal of the thymus, the clones in the blood (that originated in the thymus) persisted for at least one year, resulting in the incomplete success of the thymectomy procedure.
2. Why do MG patients treated with Rituximab relapse after a period of remission?
Another MG subtype (MuSK MG) is treated with a drug called Rituximab (RTX – an antibody that binds to CD20 on the membranes of B cells and is used to treat some autoimmune diseases and cancers), which leads to a complete stable remission of the disease phenotype, but eventually many relapse into a severe disease crisis. With the MuSK MG subtype, researchers in the O’Connor lab already knew that there is no thymus involvement and they suspected that short-lived antibody-producing cells in the circulation are the major contributors. They wanted to define the mechanism further; specifically, they wanted to know -– when patients relapse, are the autoantibody-producing clones they find entirely new or historic? They answered this question using full-length bulk BCR sequencing, patient-derived mAbs, and single-cell RNA sequencing.
Firstly, in three patients who were monitored pre-Rituximab treatment through to relapse post-Rituximab treatment, they sought persistent clones that escape RTX depletion. They did this in much the same way they did in the above thymectomy study using high-depth, bulk sequencing of the entire B cell repertoire before and after treatment. They identified clones with a common ancestor based on their V and J gene usage as well as junction sequences and found that circulating B cell clones persisted and had resisted RTX depletion. Additionally, bulk sequencing information informed them that all the persistent B cell clones had class switched, suggesting they have a role in antigen recognition.
Because the cells suspected to be major contributors to this MG-subtype are antibody secretors, they don’t have the BCR on their surface and cannot be directly isolated with a labeled antigen. However, because they had previously developed a panel of mAbs from these three patients, they had sequence information and could use this to identify the MuSK-binding, disease-causing clones in the bulk sequencing information from the same patients.
To further complement these combined approaches, both single-cell BCR sequencing and single-cell RNA sequencing/ transcriptome analysis was carried out on post-RTX treatment circulating B cells. And, in a similar way, the bulk sequencing data was used as a database to search for overlap. This was subsequently correlated with the transcriptional signature of each cell to trace the persistent cell type, which turned out to be short-lived antibody-secreting plasmablasts.
These studies illustrate the potential to gain a deeper level of understanding of the specificity of B and T cell roles in autoimmunity.
Another recent study, again sequencing BCRs, analyzed a question on everyone’s mind – How is our immune system responding to SARS-CoV-2 mRNA vaccines?
Using full-length bulk immune cell sequencing to measure the immune response to vaccination
In one of the first studies to evaluate the durability of the response to the SARS-CoV-2 mRNA vaccines, the Ellebedy lab at the Washington University School of Medicine took a closer look at the response in germinal centers (GC) in the lymph nodes of individuals who had received two doses of the Pfizer vaccine. GCs are temporary structures that form in lymphoid tissue after vaccination or infection – it’s where B cells are activated and undergo somatic hypermutation to become higher affinity, while low-affinity clones are removed. A persistent presence of GCs suggests a strong response to a vaccine. Prior to this study, the longevity of the immune response to the SARS-CoV-2 mRNA vaccines was based on whether vaccinated individuals contracted COVID-19, or by monitoring antibody levels in the blood of vaccinated individuals. So, this study takes a more intricate look at the immune response at a sequence and cellular level.
Researchers in the Ellebedy lab initially used an ELISpot assay to quantitate spike-binding IgG and IgA secreting B cells (plasmablasts) that migrated from the GC from blood taken pre- and post- immunization to identify the period of B cell response. Then, armpit lymph node aspirates showed that GCs persisted four months after the first dose of the Pfizer vaccine. The GCs contained cells with antibody-producing B cells that targeted the SARS-CoV-2 spike protein. They employed the use of mAbs (generated from GC B cells to SARS-CoV-2) and bulk B cell sequencing to examine clonal diversity in the GCs. Clonal relatives of the S-binding GC mAbs were identified among the sequences from bulk repertoire analysis of plasmablasts from PBMCs (Human Peripheral Blood Mononuclear Cells) and GC B cells. These sequences had significantly increased mutation frequencies in their variable region, indicating a vaccine-induced B cell proliferation for producing memory B cells.
While this is still a work in progress, the researchers were able to characterize the clonal diversity involved in this persistent, robust GC response, and provide insights regarding the immune response and longevity.
The new bulk cell sequencing workflow has been expertly wielded by investigators, complementing their study designs, to trace persistent cell types in Myasthenia gravis patients and to reveal aspects of SARS-CoV-2 mRNA vaccine immunity over time. Given the accessibility of this method for B and T cell receptor mRNA long reads – it will be exciting to see what comes next.
Our NEBNext® Immune Sequencing Kits are available for sequencing the full-length immune repertoires of B cells and T cells in both humans and mice.
NEB will not rent, sell or otherwise transfer your data to a third party for monetary consideration. See our Privacy Policy for details. View our Community Guidelines.
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. See www.neb.com/trademarks. The use of these products may require you to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
Don’t miss out on our latest NEBinspired blog releases!
- Sign up to receive our e-newsletter
- Download your favorite feed reader and subscribe to our RSS feed
Be a part of NEBinspired! Submit your idea to have it featured in our blog.


