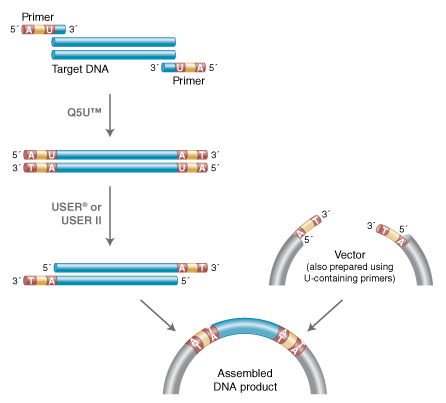
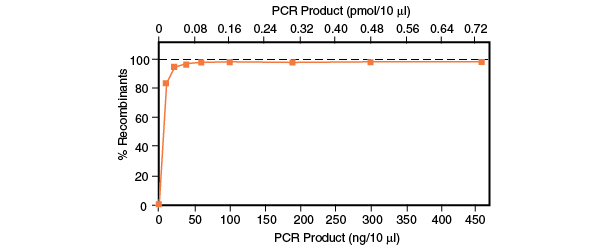
USER® Cloning
Choose Type:
- 5 Minute Transformation Protocol (C2987H/C2987I)
- 5 Minute Transformation Protocol using NEB 10-beta Competent E. coli (C3019H/C3019I)
- High Efficiency Transformation Protocol (C2987H/C2987I)
- High Efficiency Transformation Protocol using NEB 10-beta Competent E. coli (High Efficiency) (C3019H/C3019I)
- High Efficiency Transformation Protocol for 96-well format (C2987P)
- 14 Minute Transformation Protocol (NEB #C2987H/C2987I)
- High Efficiency Transformation Protocol for 384-well format (C2987R)
- High Efficiency Transformation Protocol for 96-tube format (C2987U)
- Molecular Cloning Technical Guide
- Synthetic Biology/DNA Assembly Selection Chart
Brochures
Selection Tools
- Wang S, Li W, Wang S, Hu B (2014) Rapid and Efficient Assembly of Transcription Activator-Like Effector Genes by USER Cloning J Genet Genomics; 41(6), 339-47. PubMedID: 24976123, DOI: 10.1016/j.jgg.2014.05.002
- Lund AM, Kildegaard HF, Petersen MB, Rank J, Hansen BG, Andersen MR, Mortensen UH (2014) A versatile system for USER cloning-based assembly of expression vectors for mammalian cell engineering PLoS One; 9(5), e96693. PubMedID: 24879460, DOI: 10.1371/journal.pone.0096693
- Villiers BR, Stein V, Hollfelder F (2010) USER friendly DNA recombination (USERec): a simple and flexible near homology-independent method for gene library construction Protein Eng Des Sel; 23(1), 1-8. PubMedID: 19897542, DOI: 10.1093/protein/gzp063
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.