6C. Quality Control Check and Size Selection using AMPure XP Beads NEBNext Small RNA Library Prep Set for Illumina (E7300, E7580, E7560, E7330)
This protocol follows Protocol for use with NEBNext Small RNA Library Prep Set for Illumina E7300, E7580, E7560, E7330
Note: Bead size selection is only recommended for samples showing no primer dimer and no adaptor dimer on Bioanalyzer. It will be suitable to remove peaks > 150 bp. If fragments larger than 150 bp are abundant, two rounds of bead size selection may be necessary to completely eliminate the high molecular weight fragments.
6C.1. Purify the PCR amplifed cDNA construct (100 µl) using a Monarch PCR & DNA Kit.
IMPORTANT: Use the 7:1 ratio of binding buffer:sample. Discard the flow through after each centrifugation step.
6C.2. Elute amplifed DNA in 27.5 µl Nuclease-free Water.
![]() Safe Stopping Point: It is safe to store the library at -20°C after PCR cleanup.
Safe Stopping Point: It is safe to store the library at -20°C after PCR cleanup.
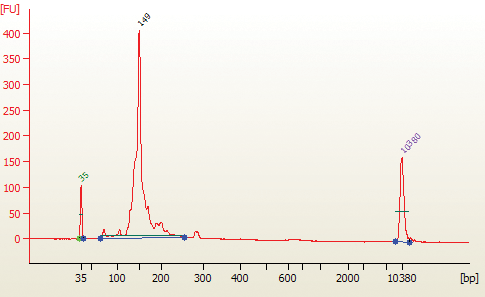
6C.3. Load 1 µl of the purified PCR reaction on the Bioanalyzer using a DNA 1000 chip according to the manufacturer's instructions (Figure 1).
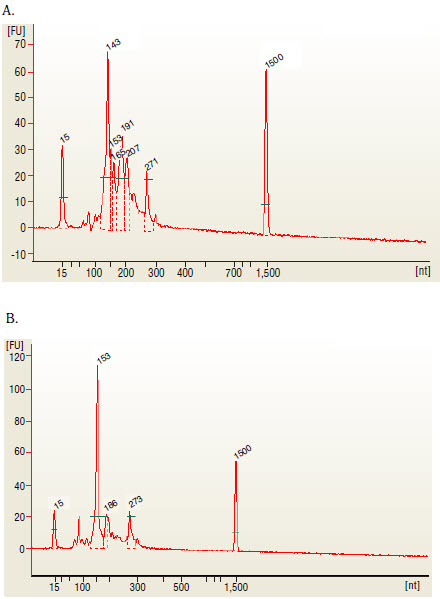
The 143 and 153 bp bands correspond to miRNAs and piRNAs, respectively. The bands on the Bionalyzer electropherograms resolve in sizes ~ 6-8 nucleotides larger than sizes observed on PAGE gels and can shift from sample to sample due to an incorrect identifcation of the marker by the bioanalyzer software. miRNA peak should be ~ 143-146 bp
6C.4. To the purified PCR reaction (25 µl), add 32.5 μl (1.3X) of resuspended AMPure XP beads and mix well on a vortex mixer or by pipetting up and down at least 10 times.
6C.5. Incubate for 5 minutes at room temperature.
6C.6. Place the tube on an appropriate magnetic stand to separate beads from supernatant. After the solution is clear (about 5 minutes), carefully transfer the supernatant (57.5 µl) to a new tube (Caution: do not discard the supernatant). Discard beads that contain the large DNA fragments.
6C.7. Add 92.5 μl (3.7X) of resuspended AMPure XP beads to the supernatant (57.5 μl), mix well and incubate for 5 minutes at room temperature.
6C.8. Place the tube on an appropriate magnetic stand to separate beads from supernatant. After the solution is clear (about 5 minutes), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets (Caution: do not discard beads).
6C.9. Add 200 μl of freshly prepared 80% ethanol to the tube while in the magnetic stand. Incubate at room temperature for 30 seconds, and then carefully remove and discard the supernatant.
6C.10. Repeat Step 6C.9. once.
6C.11. Briefly spin the tube, and put the tube back in the magnetic stand.
6C.12. Completely remove the residual ethanol, and air dry beads for up to 10 minutes while the tube is on the magnetic stand with lid open.
Caution: Do not overdry the beads, which may result in lower recovery of the DNA target. Elute the sample when the beads are still dark brown and glossy looking, but when all visible liquid has evaporated. When the beads turn lighter brown and start to crack, they are too dry.
6C.13. Elute the DNA target from the beads with 15 μl nuclease-free water. Mix well on a vortex mixer or by pipetting up and down, incubate for 2 minutes and put the tube in the magnetic stand until the solution is clear.
6C.14. Transfer the supernatant to a clean PCR tube.
6C.15. Run 1 µl on the Bioanalyzer High Sensitivity chip. Check peak distribution and concentration of the small RNA library.