6B. Quality Control Check and Size Selection using Pippin Prep NEBNext Small RNA Library Prep Set for Illumina (E7300, E7580, E7560, E7330)
This protocol follows Protocol for use with NEBNext Small RNA Library Prep Set for Illumina E7300, E7580, E7560, E7330
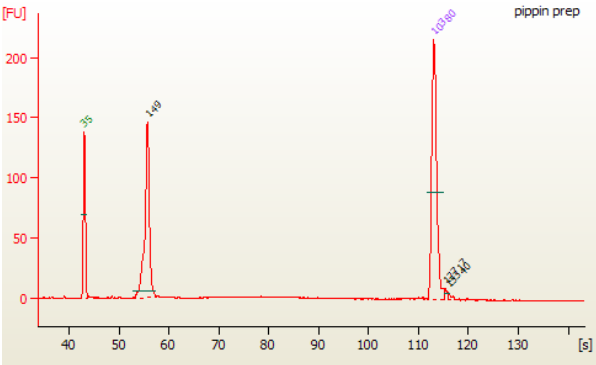
Size selection of the Small RNA library (147 bp) can done on Pippin Prep instrument using the 3% Agarose, dye free gel with internal standards (Sage Science #CDP3010).
6B.1. Purify the PCR amplifed cDNA construct (100 μl) using a Monarch PCR & DNA Cleanup Kit.
IMPORTANT: Use the 7:1 ratio of binding buffer:sample. Discard the flow through after each centrifugation step.
6B.2. Elute amplifed DNA in 32 μl nuclease-free water.
![]() Safe Stopping Point: It is safe to store the library at -20°C after PCR cleanup.
Safe Stopping Point: It is safe to store the library at -20°C after PCR cleanup.
It is recommended to QC your library before performing size selection:
6B.3. Load 1 μl of the purifed PCR reaction on the Bioanalyzer using a DNA 1000 chip according to the manufacturer's instructions (Figure 1). miRNA library should appear as a peak at 147 bp peak (that correspond for 21 nucleotide insert).
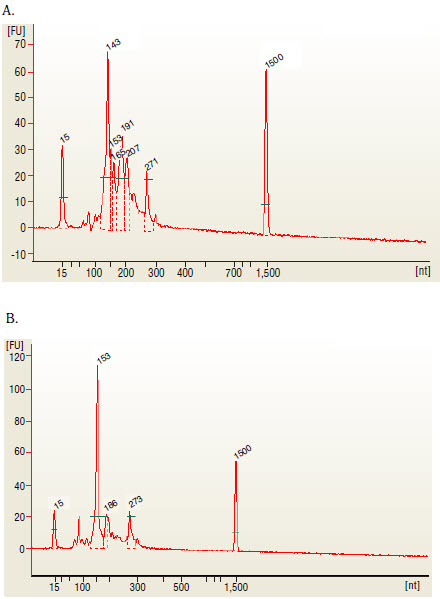
The 143 and 153 bp bands correspond to miRNAs and piRNAs, respectively. The bands on the Bionalyzer electropherograms resolve in sizes ~ 6-8 nucleotides larger than sizes observed on PAGE gels and can shift from sample to sample due to an incorrect identifcation of the marker by the bioanalyzer software. miRNA peak should be ~ 143-146 bp
Program the protocol for size selection on Pippin Prep Instrument as follows:
6B.4. In the Pippin Prep software, go to the Protocol Editor Tab.
6B.5. Click “Cassette” folder, and select “3% DF Marker P”.
6B.6. Select the collection mode as “Range” and enter the size selection parameters as follow: BP start (105) and the BP end (155). BP Range Flag should indicate “broad”.
Note: This protocol is optimized to select for 147–149 bp peak. When targeting other small RNA these settings may have to be adjusted.
6B.7. Click the “Use of Internal Standards” button.
6B.8. Make sure the “Ref Lane” values match the lane numbers.
6B.9. Press “Save As” and name and save the protocol.
Prepare sample for size selection as follows:
6B.10. Bring loading solution to room temperature.
6B.11. For each sample, combine 30 µl sample with 10 µl of DNA marker P (labeled P).
6B.12. Mix samples thoroughly (vortex mixer). Briefly centrifuge to collect.
6B.13. Load 40 µl (DNA plus marker) on one well of the 3% agarose cassette.
6B.14. Run the program with the settings indicated above.
6B.15. After sample has been eluted, collect 40 µl sample from elution well. Run 1 µl in a Bioanalzyer using the high sensitivity chip.
Note: If the Ethidium Bromide free cassettes was used, no purifcation is required before running sample on the bioanalyzer.