Types of Restriction Endonucleases
Return to Restriction EndonucleasesWhat is a Restriction Enzyme?
Restriction enzymes are traditionally classified into four types on the basis of subunit composition, cleavage position, sequence specificity and cofactor requirements. However, amino acid sequencing has uncovered extraordinary variety among restriction enzymes and revealed that at the molecular level, there are many more than four different types.
Type I Enzymes
Type I enzymes are complex, multisubunit, combination restriction-and-modification enzymes that cut DNA at random far from their recognition sequences. Originally thought to be rare, we now know from the analysis of sequenced genomes that they are common. Type I enzymes are of considerable biochemical interest, but they have little practical value since they do not produce discrete restriction fragments or distinct gel-banding patterns.
Type II Enzymes
Type II enzymes cut DNA at defined positions close to or within their recognition sequences. They produce discrete restriction fragments and distinct gel banding patterns, and they are the predominant class used in the laboratory for routine DNA analysis and gene cloning. Rather than forming a single family of related proteins, Type II enzymes are a collection of unrelated proteins of many different sorts. Type II enzymes frequently differ so completely in amino acid sequence from one another, and indeed from every other known protein, that they exemplify the class of rapidly evolving proteins that are often indicative of involvement in host-parasite interactions.
Most Common Type II Enzymes
The most common Type II enzymes are those like HhaI (NEB #R0139), HindIII (NEB #R0104), and NotI (NEB #R0189), that cleave DNA within their recognition sequences. Enzymes of this kind are the principal ones available commercially. Most recognize DNA sequences that are symmetric, because they bind to DNA as homodimers, but a few, (e.g., BbvCI (NEB #R0601): CCTCAGC) recognize asymmetric DNA sequences, because they bind as heterodimers. Some enzymes recognize continuous sequences (e.g., EcoRI (NEB #R0101): GAATTC) in which the two half-sites of the recognition sequence are adjacent, while others recognize discontinuous sequences (e.g., BglI (NEB#R0144): GCCNNNNNGGC) in which the half-sites are separated. Cleavage leaves a 3´-hydroxyl on one side of each cut and a 5´-phosphate on the other. They require only magnesium for activity and the corresponding modification enzymes require only S-adenosylmethionine. They tend to be small, with subunits in the 200-350 amino acid range.
Type IIS Enzymes
Type IIG Enzymes
Type IIG restriction enzymes, the third major kind of Type II enzyme, are large, combination restriction-and-modification enzymes, 850-1250 amino acids in length, in which the two enzymatic activities reside in the same protein chain. These enzymes cleave outside of their recognition sequences and can be classified as those that recognize continuous sequences (e.g., AcuI (NEB #R0641): CTGAAG) and cleave on just one side; and those that recognize discontinuous sequences (e.g., BcgI (NEB #R0545): CGANNNNNNTGC) and cleave on both sides releasing a small fragment containing the recognition sequence. The amino acid sequences of these enzymes are varied, but their organization is consistent. They comprise an N-terminal DNA-cleavage domain joined to a DNA-modification domain and one or two DNA sequence-specificity domains forming the C-terminus or present as a separate subunit. When these enzymes bind to their substrates, they switch into either restriction mode to cleave the DNA, or modification mode to methylate it.
Type III Enzymes
Type III enzymes are also large combination restriction-and-modification enzymes. They cleave outside of their recognition sequences and require two such sequences in opposite orientations within the same DNA molecule to accomplish cleavage; they rarely give complete digests.
Type IV Enzymes
Type IV enzymes recognize modified, typically methylated DNA and are exemplified by the McrBC and Mrr systems of E. coli.
Videos
-
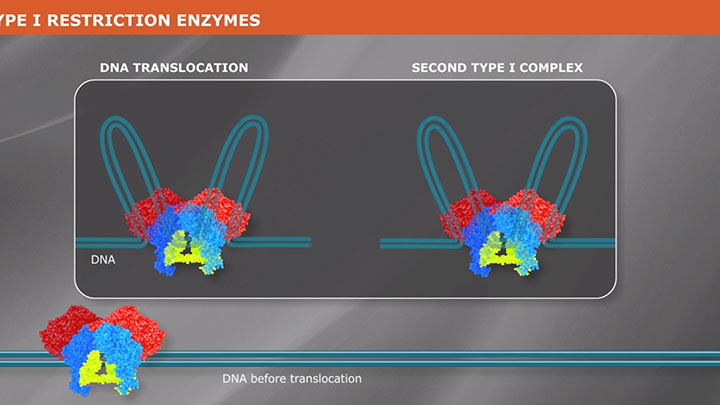
What is a Type I Restriction Enzyme?
Type I restriction enzymes are a group of endonucleases that recognize a bipartite sequence, but do not produce a predictable cleavage pattern. Learn more about how Type I REs work.
-
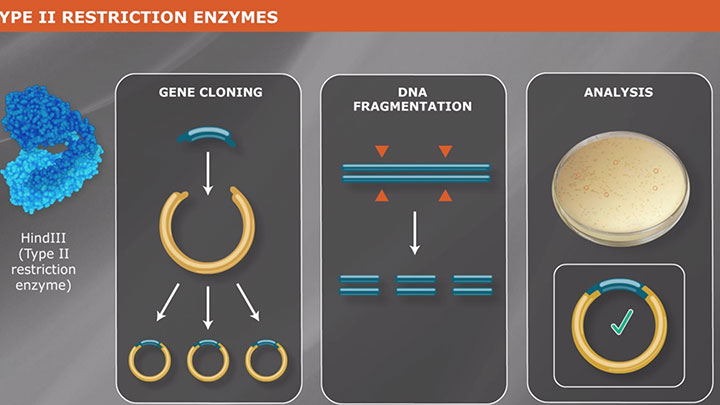
What is a Type II Restriction Enzyme?
Type II restriction enzymes are most commonly used for molecular biology applications, as they recognize stereotypical sequences and produce a predictable cleavage pattern. Learn more about how Type II REs work.
-
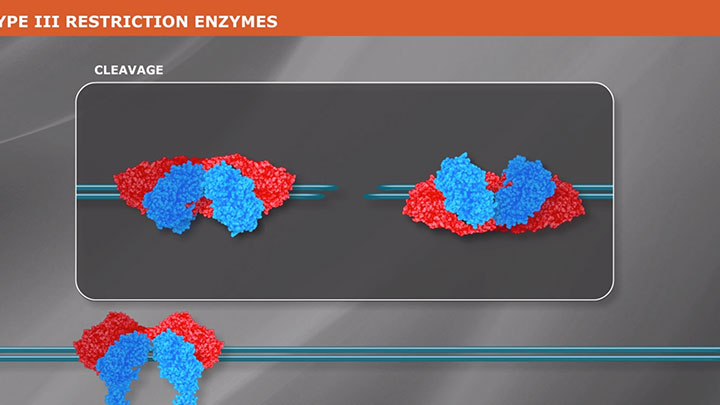
What is a Type III Restriction Enzyme?
Type III restriction enzymes are a group of endonucleases that recognize a non-pallindromic sequence, comprising two inversely oriented sites. Learn more about these poorly understood enzymes.

