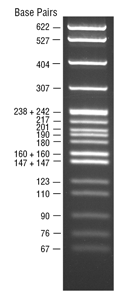
pBR322 DNA-MspI Digest
The large size of this product was discontinued on 12/31/2020. The small size will continue to be available.
- Size range: 9 bp - 622 bp
- Supplied with free vial of Gel Loading Dye, Purple (6X), no SDS (NEB #B7025)
- Small size suitable for 50 gel lanes
Featured Video
-
Product Information
The MspI digest of pBR322 DNA yields 26 fragments. Suitable for use as molecular weight standards for agarose gel electrophoresis (1,2). Comes supplied with 1 vial of Gel Loading Dye, Purple (6X), no SDS.
- Recommended gel percentage range: 2-3.5%
- Optimum separation on 3%
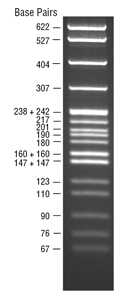
pBR322 DNA-MspI Digest visualized by ethidium bromide staining on a 3% agarose gel.
Suggested Load: 1 µg/ gel lane.- This product is related to the following categories:
- DNA Markers & Ladders Products
- This product can be used in the following applications:
- DNA Analysis
-
Protocols, Manuals & Usage
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.