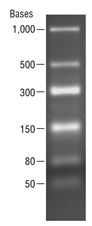
Low Range ssRNA Ladder
- Size range: 50 bases - 1,000 bases
- Compatible with denaturing polyacrylamide-urea gels, denaturing agarose gels and native agarose gels
- Convenient reference band at 300 bases
Featured Video
-
Product Information
The Low Range ssRNA Ladder is a set of 6 RNA molecules produced by in vitro transcription of a mixture of 6 linear DNA templates. The ladder sizes are: 1000, 500, 300, 150, 80 and 50 bases. The 300 base fragment is at double intensity to serve as a reference band. This ladder is suitable for use as an ssRNA size standard on denaturing polyacrylamide-urea gels, and on denaturing or native agarose gels.
Usage Recommendation
This marker was not designed for precise quantification of ssRNA mass.
Denaturing vs. Native Agarose Gels
It is common practice to electrophorese RNA on a fully denaturing agarose gel, such as one containing formaldehyde (1). However, in many cases it is possible to run RNA on a native agarose gel and obtain suitable results. In fact, it has been demonstrated that treatment of RNA samples in a denaturing buffer maintains the RNA molecules in a denatured state, during electrophoresis, for at least 3 hours (2,3). The use of native agarose gels eliminates problems associated with toxic chemicals and the difficulties encountered when staining and blotting formaldehyde gels.
Sample Preparation
The Low Range ssRNA Ladder is also compatible with formaldehydebased loading buffers.
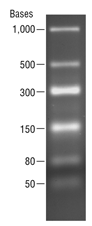
1 µg/lane. 2.0% TBE agarose gel. - This product is related to the following categories:
- RNA Markers & Ladders Products,
- RNA Markers and Ladders
-
Protocols, Manuals & Usage
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.