T3 DNA Ligase
T3 DNA Ligase will ligate these substrates:
dsDNA
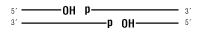
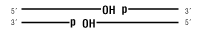
Nicked DNA/RNA

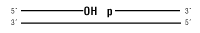
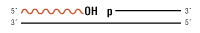

1. Note: Trace amounts of this substrate can be ligated with overnight incubation, and ligation is further enhanced in Stick Together DNA Ligase Buffer. Please note that Blunt TA Ligase Mastermix is the recommended ligase product for this substrate.
- T3 DNA Ligase is an ATP-dependent dsDNA Ligase from bacteriophage T3
- 2x salt tolerence as compared to T4 DNA Ligase
- This product can be purchased in larger volumes. Submit an inquiry to find out more.
- Not sure which ligase to choose? Refer to our DNA and RNA Ligase Properties Chart or DNA Ligase Selection Chart
Featured Video
-
Product Information
T3 DNA Ligase is an ATP-dependent ds DNA ligase from bacteriophage T3. It will catalyze the formation of a phosphodiester bond between adjacent 5´ phosphate and 3´ hydroxyl groups of duplex DNA. Cohesive ends, blunt ends, and nick sealing can all be efficiently catalyzed by T3 DNA Ligase (1). As with T4 DNA Ligase, blunt end ligation is enhanced by the addition of PEG 6000 to the reaction (2). T3 DNA Ligase exhibits a higher tolerance (2-fold) for NaCl in the reaction compared to T4 DNA Ligase, making the enzyme a versatile choice for in vitro molecular biology protocols requiring DNA ligase activity. Product Source
An E. coli strain containing a recombinant gene encoding T3 DNA Ligase.- This product is related to the following categories:
- DNA Ligases Products
- This product can be used in the following applications:
- Cloning Ligation
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.