EpiMark® 5-hmC and 5-mC Analysis Kit
- Catalog # E3317 was discontinued on December 18, 2023
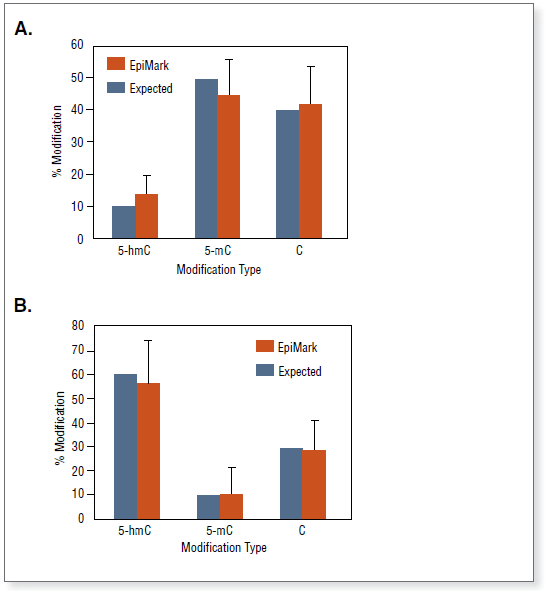
The EpiMark® 5-hmC and 5-mC Analysis Kit can be used to analyze and quantitate 5-methylcytosine and 5-hydroxymethylcytosine within a specific locus. Complete conversion of 5-hmC to glucosylated 5-hmC in DNA by T4 β-glucosyltransferase (T4-BGT).
- Discrimination between 5-mC and 5-hmC in CCGG sequences using enzymatic digestion and PCR amplification
- Relative quantitation of 5-mC and 5-hmC
- Easy-to-use protocol
- This kit contains enough material for 20 reactions

-
Product Information
5-methylcytosine (5-mC) is the predominant epigenetic mark in mammalian genomic DNA. 5-hydroxymethylcytosine (5-hmC) is a newly discovered epigenetic modification that is presumably generated by oxidation of 5-mC by the TET family of cytosine oxygenases.1,2
Techniques exist that can identify 5-mC in genomic DNA, but the most commonly used method, bisulfite sequencing, is laborious and cannot distinguish between 5-mC from 5-hmC.3
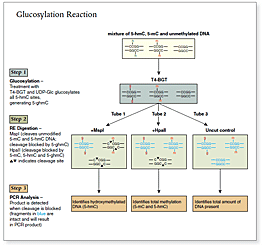
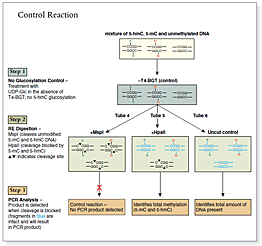
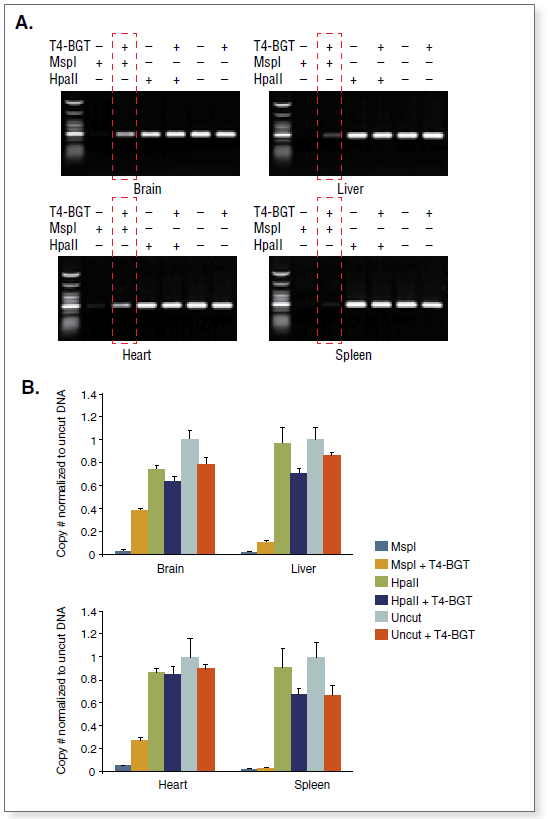
The kit distinguishes 5-mC from 5-hmC by the addition of glucose to the hydroxyl group of 5-hmC via an enzymatic reaction utilizing T4 β-glucosyltransferase (T4-BGT). When 5-hmC occurs in the context of CCGG, this modification converts a cleavable MspI site to a noncleavable one.
An overview of the detection procedure is summarized in Figure 1.
Control DNA Sequence
5´-CAGTGAAGTTGGCAGACTGAGCCAGGTCCCACAGATGCAGTGACCGGAGT
CATTGCCAAACTCTGCAGGAGAGCAAGGGCTGTCTATAGGTGGCAAGTCA-3´
Control DNA substrates are synthetic 100 bp double stranded fragments containing a single MspI/HpaII site (CCGG). The three fragments are identical except for modification of the internal C in this site.
FW Primer Sequence
5´- CA GTG AAG TTG GCA GAC TGA GC -3´
REV Primer Sequence
5´- CTG ACT TGC CAC CTA TAG ACA GC -3´- This product is related to the following categories:
- Discontinued (<3 years)
-
Protocols, Manuals & Usage
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.