O-Glycosidase & Neuraminidase Bundle
This bundle contains 1 vial each of O-Glycosidase (NEB #P0733S) and a2-3,6,8 Neuraminidase (NEB #P0720S), which can be used simultaneously for the removal of terminal sialic acid residues and core 1 and core 3 O-glycans.
- Recombinant enzymes with no detectable endoglycosidase or other exoglycosidase contaminating activities
- Glycerol-free for optimal performance in HPLC and mass spectrometry analysis
- ≥95% purity, as determined by SDS-PAGE and intact ESI-MS
- Optimal activity and stability for up to 24 months

-
Product Information
1 set of this bundle includes 2,000,000 units of O-Glycosidase and 2,000 units of Neuraminidase.
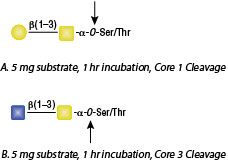
O-Glycosidase, also known as Endo-α-N-Acetylgalactosaminidase, catalyzes the removal of Core 1 and Core 3 O-linked disaccharides from glycoproteins.
Neuraminidase is the common name for Acetyl-neuraminyl hydrolase (Sialidase). This Neuraminidase catalyzes the hydrolysis of α2-3, α2-6, and α2-8 linked N-acetyl-neuraminic acid residues from glycoproteins and oligosaccharides.
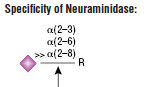
Product Source
O-Glycosidase is cloned from Enterococcus faecalis and expressed in E.coli (1). Neuraminidase is cloned from Clostridium perfringens (2) and overexpressed in E. coli (3).- This product is related to the following categories:
- Endoglycosidases Products
- This product can be used in the following applications:
- Expression Systems,
- Protein Digestion,
- Glycan Sequencing,
- Proteomics,
- Recombinant Glycoprotein Expression, Glycoprotein Analysis
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.