Adding Capabilities to CRISPR-Cas-Based Biotechnology
Posted on Tuesday, May 30, 2023
By
Topic: What is Trending in Science
The Robb Lab at NEB collaborates with both academic and industry researchers in their mission of discovery, characterization, and application development efforts with CRISPR-Cas systems. It’s always galvanizing to look to the origin of CRISPR tools and consider how this scientific field has expanded into multiple substrate types and applications over time. While NEB produces many useful Cas nucleases to support research and development, the Robb Lab is always pursuing novel, distinctive properties – inspired by the abundance and diversity of Cas nucleases found in nature.
Phages and plasmids beware
You’ve got to hand it to evolution. CRISPR systems are a marvel. Bacteria and archaea with CRISPR systems can rapidly target foreign plasmids or phages for destruction, kick off other immune responses and pass that playbook on to cell progeny.
Here’s the premise of CRISPR. Pieces of the invader’s DNA are recorded as spacer sequences among repeat sequences within the CRISPR array of a bacterial or archaeal chromosome. These spacer sequences are acquired by CRISPR-associated (Cas) nuclease protein modules intercepting foreign sequences. Effector genes, such as the Cas nucleases themselves, are adjacent to the CRISPR array – if these genes aren’t encoded elsewhere in the genome. Spacer sequences in a CRISPR array are transcribed in a single transcript and then processed to generate short RNAs or transcribed as individual short RNAs. Fittingly, these short RNAs are known as CRISPR RNAs (crRNAs). A subset of CRISPR-Cas systems also requires a second encoded small non-coding trans-activating CRISPR RNA (tracr) or scoutRNA, which the crRNA will hybridize. In either case, the single crRNA, or the paired crRNA-tracrRNA molecule, will complex with Cas proteins to form a ribonucleoprotein (RNP) complex. When an RNP effector complex recognizes a short sequence motif termed the Protospacer Adjacent Motif (PAM), RNA-guided Cas RNPs use the spacer sequence of the RNA to guide the cleavage of complementary strands of foreign nucleic acid sequence targets. It’s a forensic detection system and elimination control rolled into one.
Speedy retargeting makes CRISPR powerful at the lab bench
The biggest advantage of using CRISPR is its retargeting efficiency. That’s what propelled it into the world of gene editing from bacterial adaptive immunity research.
Before the application of CRISPR, most precision gene editing methods employed zinc finger or TAL effector nucleases. These systems rely on protein-binding domains to target DNA. Once it was established that Cas9 nuclease cut invading DNA within a complex guided by an RNA complementary to the target DNA sequence, the gene editing potential was realized by the research community. Genome editing with Cas9 was simplified from a three-component into a two-component system that combined trRNA and crRNA into a single synthetic single guide RNA (sgRNA). Applying RNA-guided Cas9 nuclease to eukaryotic cells in the presence of a donor DNA template triggered the homology-directed repair pathway to make precise mutations at targeted sites. Without any donor template, the non-homologous end-joining repair pathway results in insertions and/or deletions to disrupt the target.
Observations of native bacterial CRISPR systems and a decade of scientific explorations into the capabilities of natural and engineered nucleases for gene editing coalesced to inspire CRISPR/Cas-based gene editing. Delivery formats expanded from plasmid-based to viral delivery, mRNA and direct delivery of the ribonucleoprotein (RNP) complex. Genome editing remains the biggest use of CRISPR-Cas nucleases in research today.
Easy and rapid retargeting of Cas9 nuclease for genome editing with single guide RNA introduced unparalleled workflow efficiency. In contrast, it takes extensive effort to re-target alternate gene editing systems based on fusion protein engineering or meganuclease selection, since a new protein variant needs to be identified for each target sequence. Ultimately, speedy retargeting with CRISPR has lowered costs for gene editing and many other goals.
Leveraging CRISPR in gene editing, pathogen detection, sequencing, and more
Tools based on wild-type characteristics and engineered enhancements of CRISPR-Cas nuclease systems have been enthusiastically adopted for a wide range of goals. Harnessing the discrete targeting and activation properties of various CRISPR/Cas complexes is a strong research focus. New capabilities are quickly being added to current platforms and new approaches are being rapidly developed.
The abundance and diversity of wild-type Cas endonucleases has only recently been fully appreciated and analyzed. Formal classification is still a work in progress, but three major classes of CRISPR systems are distinguished by multiple subunit or single Cas nuclease protein unit effector complexes. The single subunit nature of Class II, which includes Cas9, Cas12 and Cas13, makes it a workflow favorite. There are six types of Class II so far, further divided into many effector protein subtypes, with extreme divergence. As more Cas nucleases are characterized, their precise features are leveraged in workflows. Their distinct attributes include recognition targets, cleavage, overhangs on cleavage products, guide RNA sizes, PAM motifs and activity at different temperatures.
The target substrates of CRISPR-Cas nucleases factor strongly into applications. While the premise of our CRISPR origin story featured the capability to target dsDNA, target options also include single-stranded DNA (ssDNA) or RNA, as well as non-nucleic acids, like proteins. Precise CRISPR-Cas targeting of dsDNA is used for gene editing, epigenome editing, base editing, gene modulation, genome-wide functional screening, localization studies, and in vitro DNA digests. CRISPR-Cas systems with ssRNA recognition can be used in both RNA-based nucleic acid detection and RNA base editing. Certain Cas complexes that target dsDNA can also act on ssDNA. CRISPR strategies leverage precise substrate targeting alongside distinct capabilities of Cas nuclease orthologs and engineered variants to cleave, mutate, nick, unwind and stably bind to DNA and RNA.
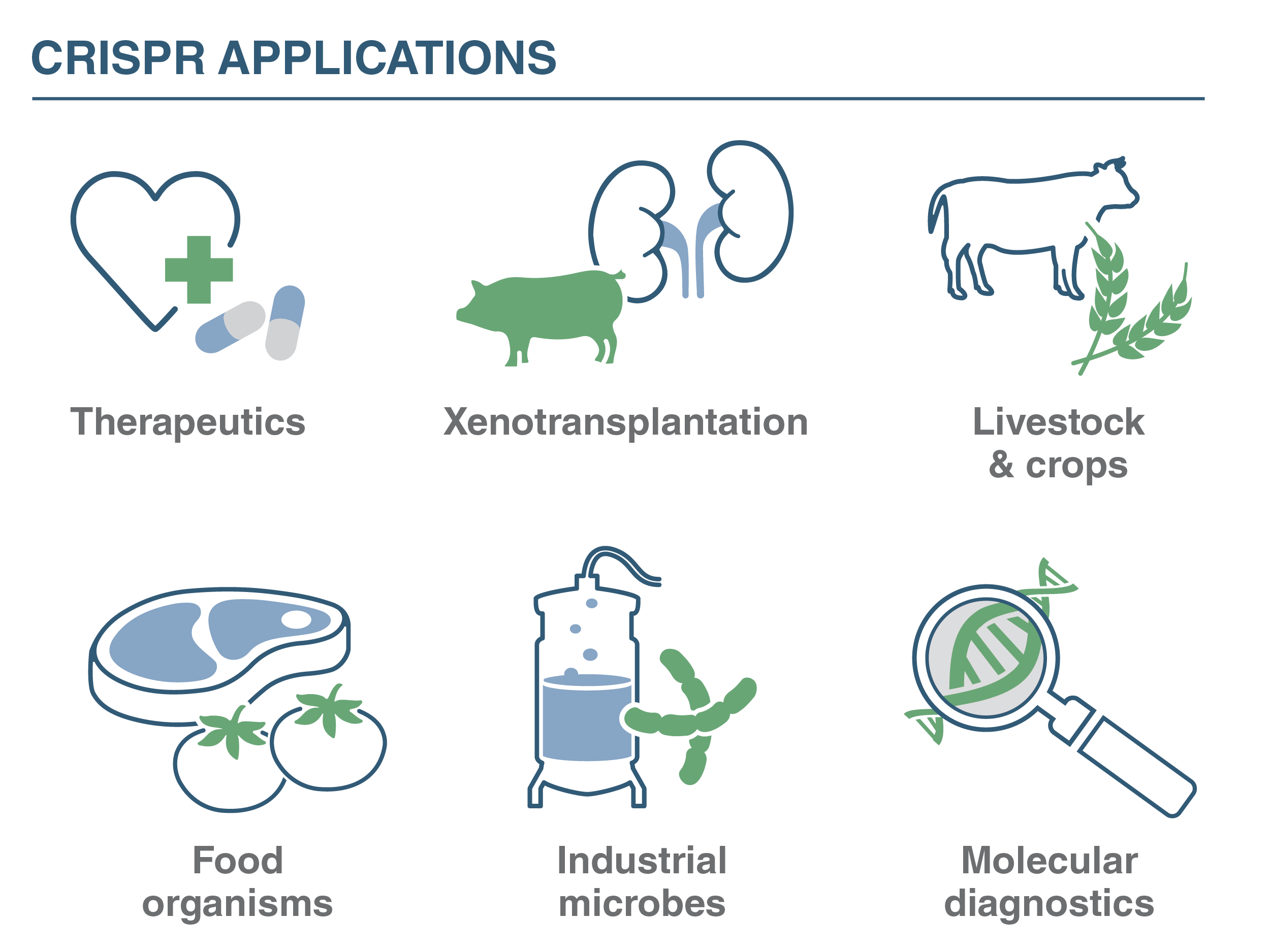
Research to remove CRISPR limitations
Scientists in The Robb Lab at New England Biolabs take part in a wide range of external collaborations with talented academic and industrial research teams. Scientific director and principal investigator, Brett Robb, Ph.D., explained that “Research into the discovery, characterization, and applications of CRISPR-Cas systems with our collaborators over the years has expanded our toolbox of enzymes and enabled exciting new applications of CRISPR-Cas proteins. In addition to being incredibly scientifically rewarding, these interactions have been inspirational and fun.” Here are a few examples.
The Robb lab supported a recent study led by The New York Genome Center’s Sanjana Lab, alongside scientists from the Synthego® corporation, demonstrating that Cas13 RNPs can successfully target mRNAs in human T-cells. Further, chemical modification of the guide RNAs improved the efficiency and duration of mRNA silencing by Cas13. The study was the first proof that efficient transcriptome engineering is possible with CRISPR-Cas13 RNPs in primary human cells. This work hints at a future where CRISPR-Cas13 gene expression modulation technology might be used to optimize cell and immune therapy outcomes for the treatment of cancers.
with our collaborators over the years has expanded our toolbox of enzymes and enabled
exciting new applications of CRISPR-Cas proteins. In addition to being incredibly
scientifically rewarding, these interactions have been inspirational and fun.
Barriers to sustainable biotechnology were lowered in a study by researchers from Wageningen University who the Robb Lab collaborated with. Bio-based production with commercial scale microbial fermentation is environmentally friendlier than fossil fuel-based industrial production, but its higher costs are problematic. Thermophilic microbe production hosts are an attractive solution. Their use can lower operation costs, raise production rates, provide higher substrate and product solubility, and carry a reduced risk of enzyme product inhibition. With these advantages in mind, Wageningen University microbiologists from the Van der Oost Lab collaborated with the Robb Lab in their work to develop a Cas12a-based genome editing tool for moderate thermophiles. Previously, Cas12a nucleases had been typically limited for use in organisms with growth temperatures up to 37 °C. Establishing the activity of the FnCas12a ortholog at higher temperatures opens opportunities for thermophilic hosts. The new tool based on this characterization has added advantages over Cas9-based systems. Importantly, the range of organisms for CRISPR-based editing can be expanded.
The Robb lab further expanded operating temperatures for CRISPR-Cas12a applications when they reported the characterization of YmeCas12a and CmeCas12a that they identified in metagenomic sequences from a hot spring and a compost pile. Cme and Yme Cas12a are active at temperatures over 50°C and over 60°C, respectively, which may enable genome editing in thermophilic organisms and Cas12a-based detection technologies at higher temperatures.
PAM compatibility of Cas nucleases can restrict the range of CRISPR-Cas nuclease applications – particularly for genome editing and base editing. A study undertaken with Caszyme, Corteva Agriscience™, Vilnius University and Iowa State University exploring the natural diversity of Cas9 orthologs showed that the Cas9 PAM interacting domain is extraordinarily flexible and has evolved to recognize a wide variety of sequences. These Cas9 protein analyses were reported in Nature Communications, noting intriguing possibilities for new combinations of non-cross reactive CRISPR-Cas9 systems in gene editing, possibilities for temperature-controlled DNA search and modification, and expanding targetable sequence space – especially important for base-editing approaches. Many Cas orthologs were identified with different advantages than those generally assigned to Spy Cas9. The future of expanding CRISPR capabilities looks bright indeed.
The foundation to expand the CRISPR toolbox
NEB focuses on discovering unique properties of Cas nucleases, opportunities for engineered Cas enzymes, and enhancing other CRISPR system components that bolster CRISPR-based tools. CRISPR-based tools are important for constructing model systems to understand disease mechanisms. They are important for pathogen detection in clinics or wastewater samples, and they are valuable for devising lifesaving therapies and precision medical diagnostics. They can open doors for bio-based manufacturing. By diving deep into biodiversity and distinctive properties of CRISPR-Cas nucleases, The Robb Lab aims to help propel the CRISPR revolution to new heights.
New England Biolabs offers a suite of Cas nucleases for R&D, including the newly released EnGen® Spy Cas9 HF1 (NEB #M0667) which can reduce off-target effects while maintaining on-target editing efficiency.
Learn More About Programmable Nucleases
NEB will not rent, sell or otherwise transfer your data to a third party for monetary consideration. See our Privacy Policy for details. View our Community Guidelines.
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. See www.neb.com/trademarks. The use of these products may require you to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
Don’t miss out on our latest NEBinspired blog releases!
- Sign up to receive our e-newsletter
- Download your favorite feed reader and subscribe to our RSS feed
Be a part of NEBinspired! Submit your idea to have it featured in our blog.


