Protein Expression in Yeast
Choose Type:
- Transformation Protocol for K. lactis GG799 Competent Cells (C1001)
- Protein Expression using the K. lactis Protein Expression Kit - Simultaneous Expression of Multiple Proteins
- Protein Expression using the K. lactis Protein Expression Kit - Transformation of K. lactis GG799 cells
- Protein Expression using the K. lactis Protein Expression Kit - Identification of properly integrated cells
- Protein expression using the K. lactis Protein Expression Kit - Cloning a PCR fragment into pKLAC2 (E1000).
- Protein Expression using the K. lactis Protein Expression Kit - Identification of Multi-copy Integrants
- Protein Expression using the K. lactis Protein Expression Kit - Growth of strains for detection of secreted protein
- Protocol II: 1 M Tris-HCl Buffer Stock Solution (1 liter)
- Protein expression using the K. lactis Protein Expression Kit - Linearization of pKLAC2 for integrative transformation of K. lactis.
- Protocol I: Yeast Carbon Base Medium Powder Agar Medium with 5 mM acetamide solution (500 ml)
-
Avoid Common Obstacles in Protein Expression
Read how to avoid common obstacles in protein expression that prevent interactions with cellular machinery.
-
Why Choose the K. lactis Protein Expression Kit?
Review the advantages of the K. lactis Protein Expression Kit for rapid, high yield protein expression in yeast.
- Protein Expression & Purification Brochure
- Protein Expression and Purification Selection Chart
Feature Articles
Brochures
Selection Tools
- Sakhtah, H., Behler, J., Ali-Reynolds, A., Causey, T.B., Vainauskas, S., Taron, C.H. (2019) A novel regulated hybrid promoter that permits autoinduction of heterologous protein expression in Kluyveromyces lactos Appl Environ Microbiol; pii: e00542-19. PubMedID: 31053583
- Chuzel, L., Ganatra, M.B., Schermerhorn, K.M., Gardner, A.F., Anton, B.P., Taron, C.H. (2017) Complete genome sequence of Kluyveromyces lactis strain GG799, a common yeast host for heterologous protein expression Genome Announc; 5(30), PubMedID: 28751387
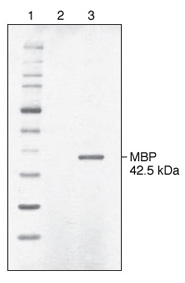
Lane 2: spent culture medium (15 µl) from wild-type K. lactis cells.
Lane 3: spent culture medium (15 µl) from K. lactis cells harboring an integrated expression cassette containing the E. coli malE gene
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.

