NEBNext® ARTIC products for SARS-CoV-2 sequencing
Choose Type:
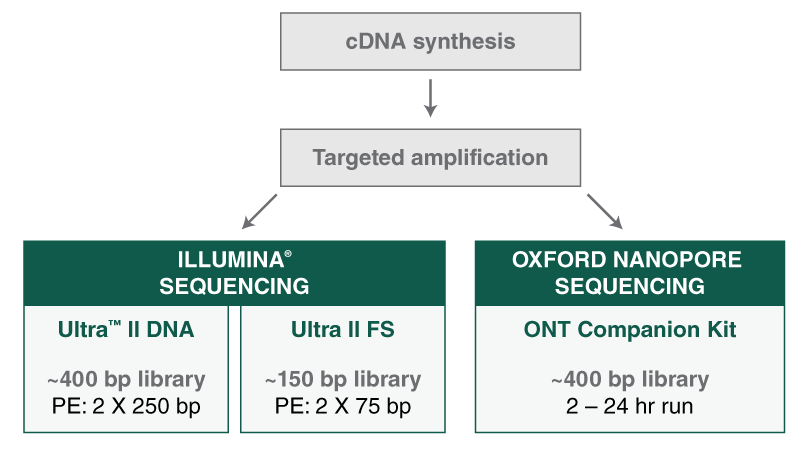
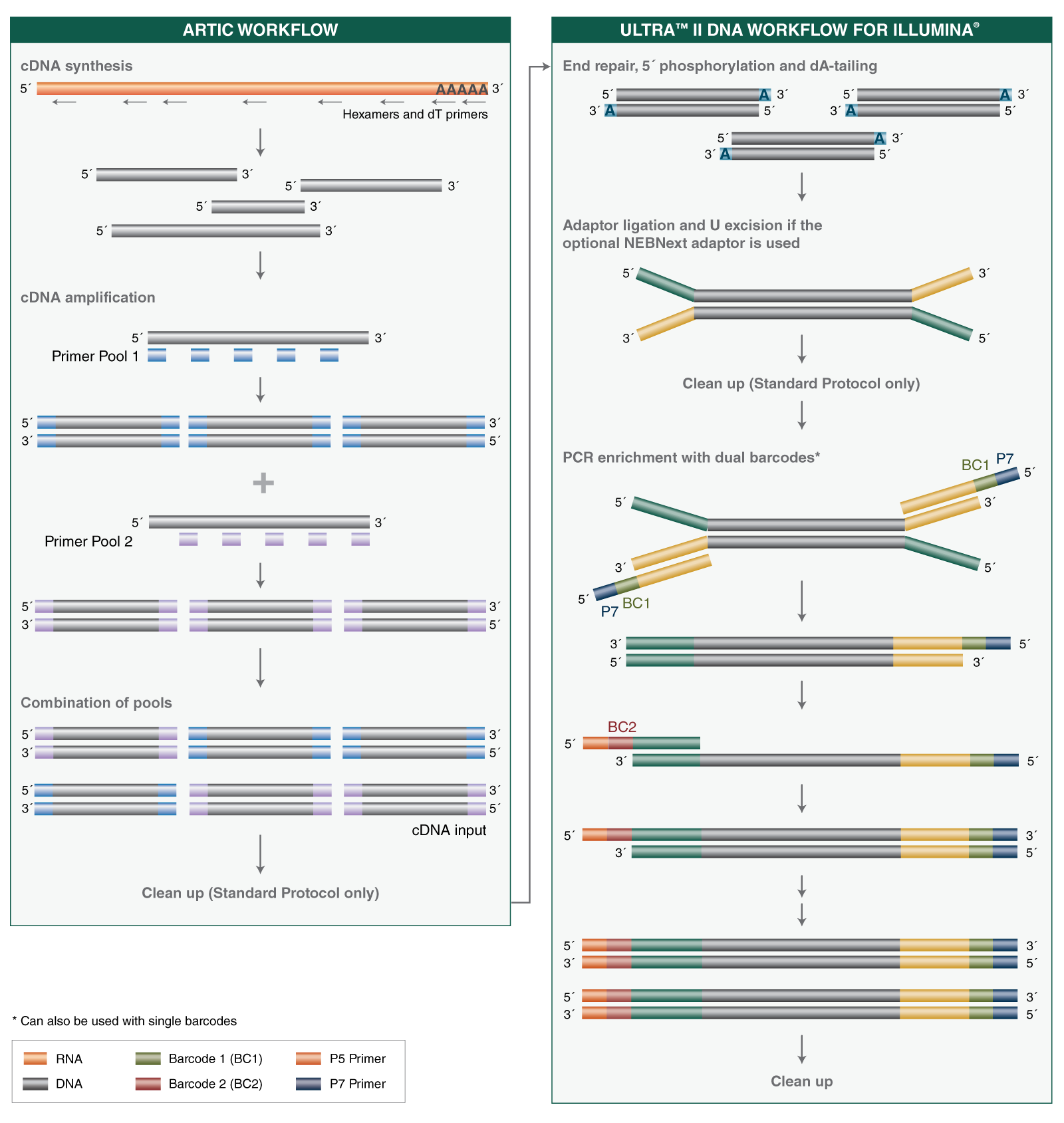
- Where can I find guidelines and protocols for using the NEBNext ARTIC SARS-CoV-2 Library Prep Kit, including cDNA synthesis, cDNA amplification and library prep?
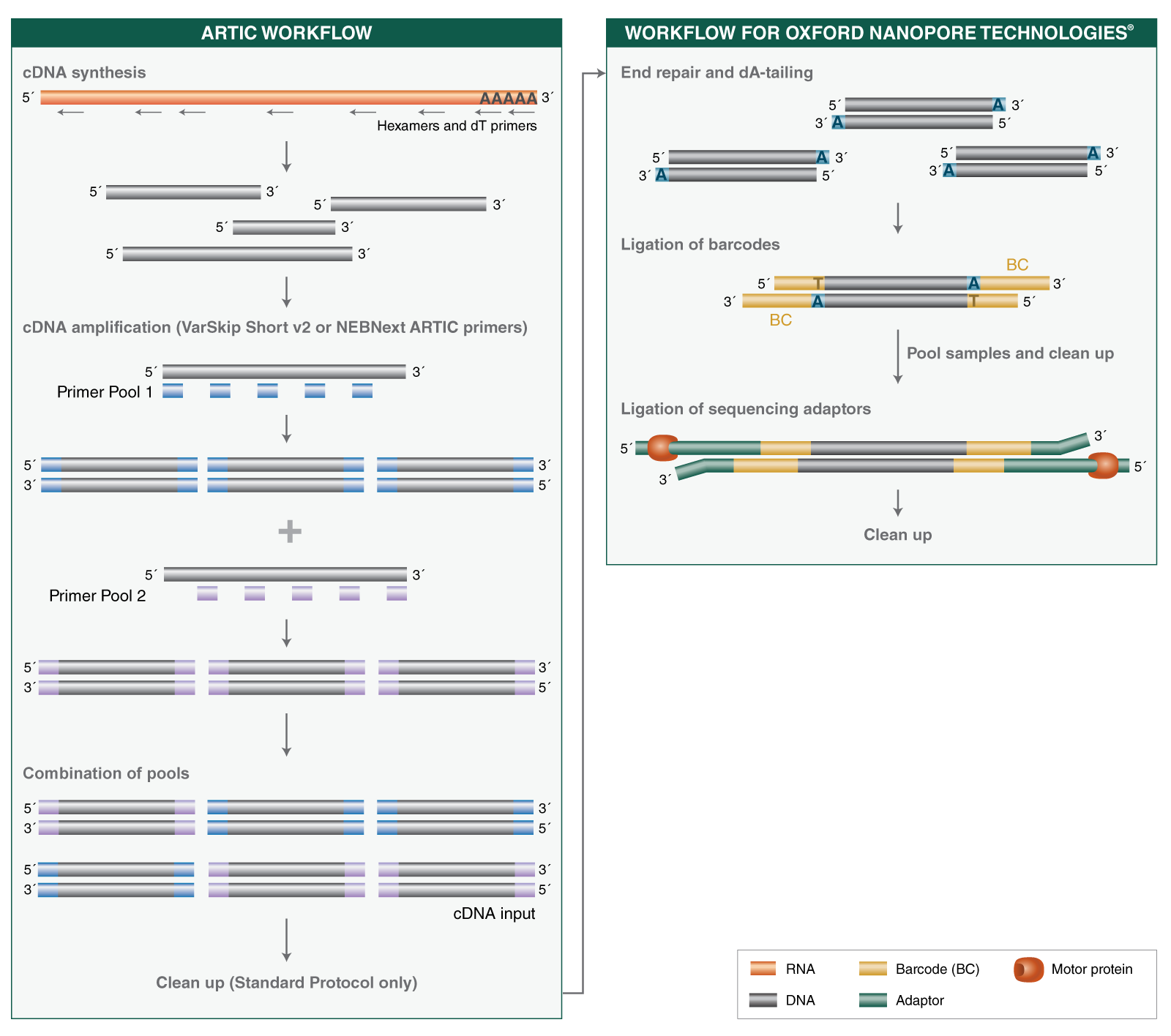
- Where can I find guidelines and protocols for using the NEBNext ARTIC SARS-CoV-2 RT-PCR Module, including cDNA synthesis and cDNA amplification?
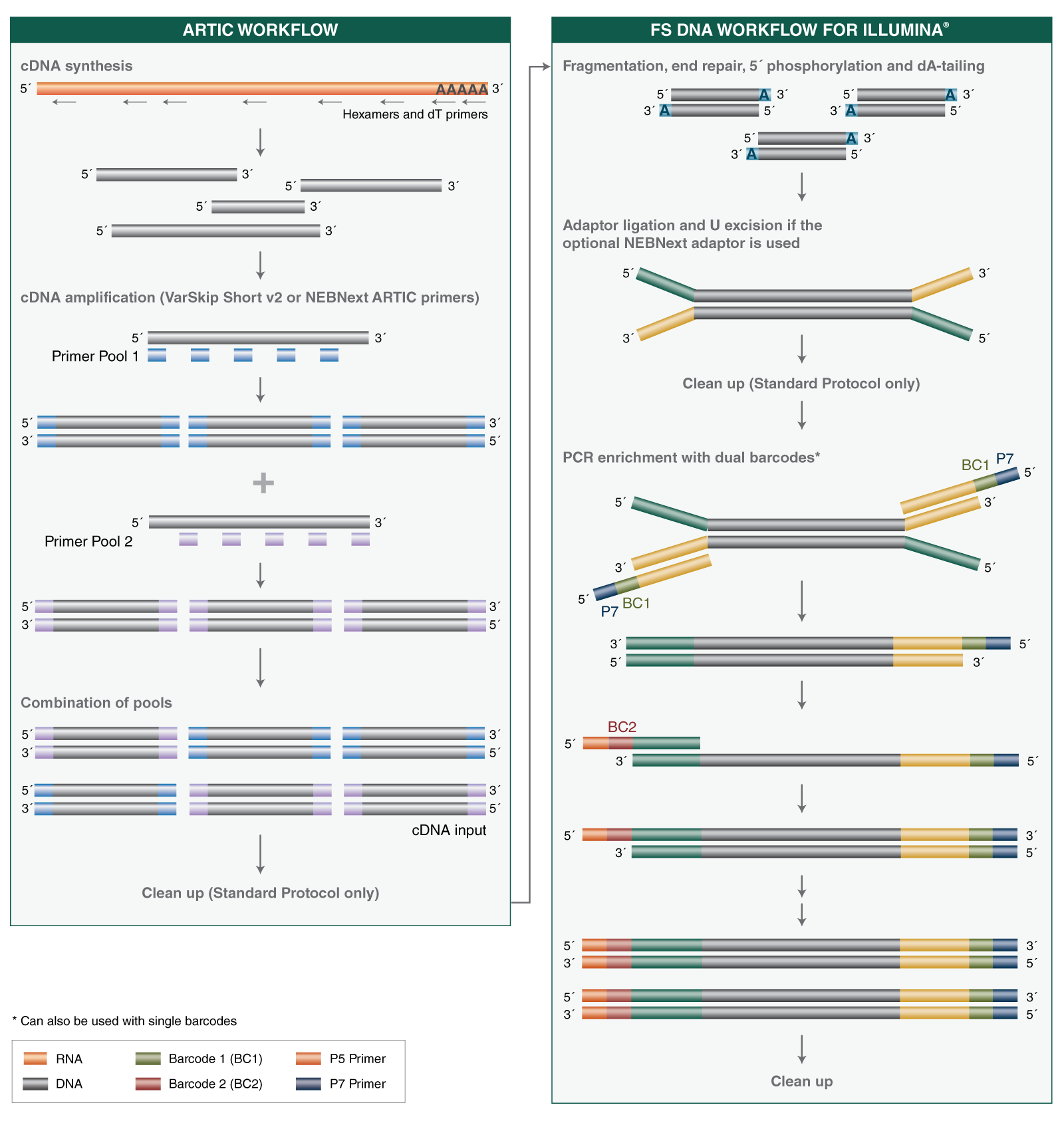
- Where can I find guidelines and protocols for using the NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit, including cDNA synthesis, cDNA amplification and library prep, and the Express protocol option with reduced cleanup steps?
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
Read what leaders in ARTIC SARS-CoV-2 sequencing are saying
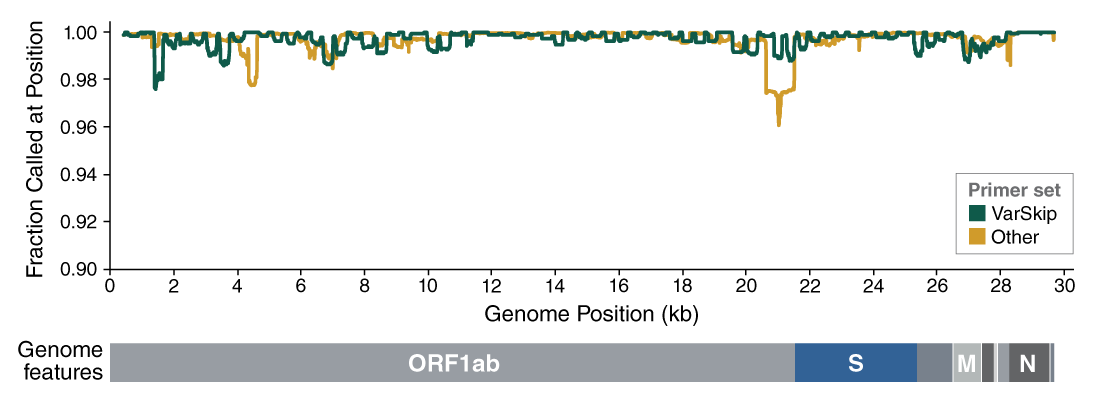
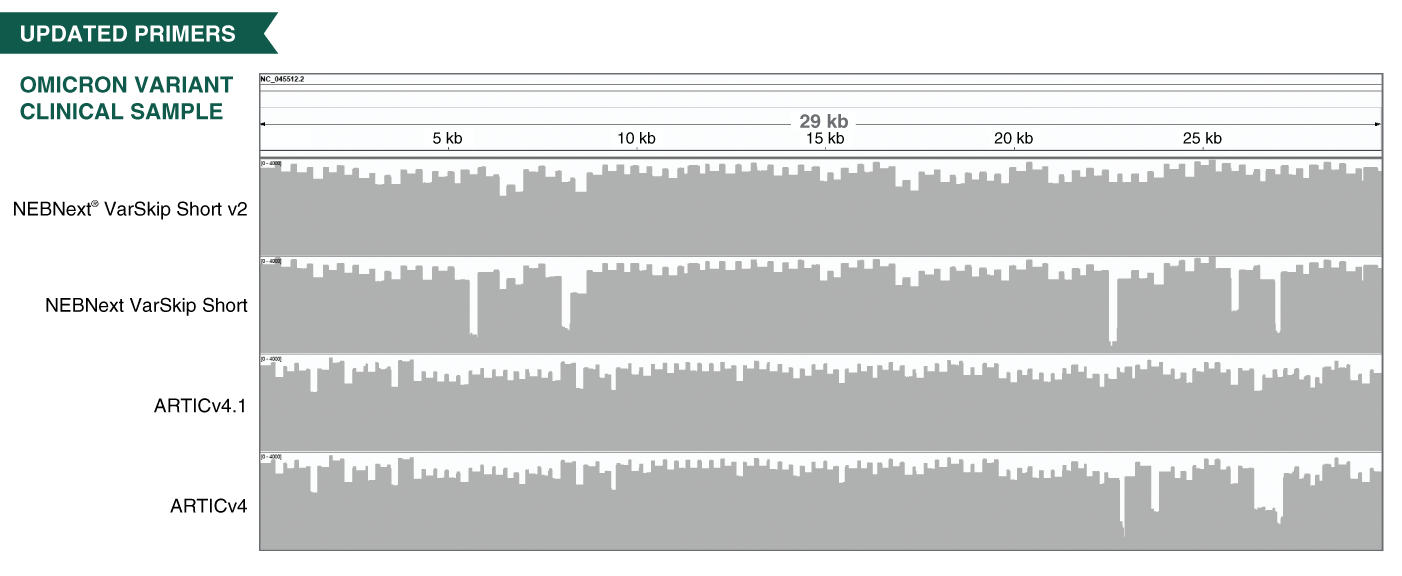
The NEBNext ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies) will provide researchers with the most convenient way to perform nanopore sequencing of SARS-CoV-2. When developing the ARTIC protocol we have focused on recovery of near-complete genomes from challenging clinical samples with <Ct33. We adopted a 400 bp tiling amplicon approach because it provides robust performance across a wide range of inputs including high Ct and degraded RNA samples which are often encountered. The native barcoding workflow provides full-length amplicons compatible with best practice demultiplexing and reference-based consensus building while the PCR-free library preparation minimizes amplicon contamination risk. We have worked in collaboration with NEB in optimizing the protocol to minimize reagent volumes while improving performance and this now also benefits from optimized primer pools which greatly improve genome completeness and reduce read coverage requirements. The compatibility with the 96-barcode native barcoding expansion kit allows the flexibility to run between 24 and 96 samples depending on your requirements.
We have been impressed by the great performance of the NEBNext ARTIC FS kit in our sequencing of human samples containing SARS-CoV-2 RNA. With this kit we have been successfully processing samples of varying quality and quantity and have been amazed by its sensitivity and remarkable uniformity of the viral genome coverage. The quick turnaround time of the robust workflow has been truly beneficial for us.
The NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit ensures rapid, accurate and reliable determination of the SARS-CoV-2 whole genome. Due to its ease of use, the NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit enables rapid integration into the routine NGS workflow. Furthermore, the kit still works robust with difficult clinical samples (ct>30). I would recommend the NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit 100%.